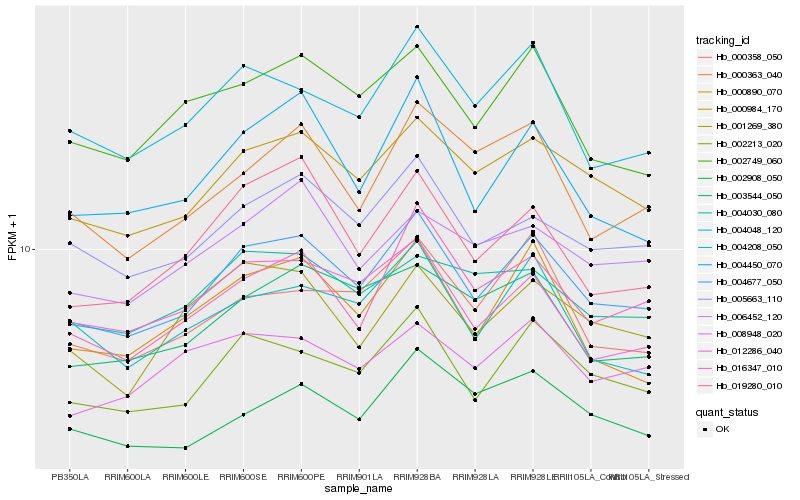
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004677_050 |
0.0 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002213_020 |
0.044575822 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000984_170 |
0.0454573811 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 4 |
Hb_005663_110 |
0.0580170865 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_016347_010 |
0.0630267917 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 6 |
Hb_004208_050 |
0.0645916228 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 7 |
Hb_002749_060 |
0.0675549175 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 8 |
Hb_004450_070 |
0.0676954795 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 9 |
Hb_012286_040 |
0.0706265222 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 10 |
Hb_000890_070 |
0.0717673134 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004048_120 |
0.0728544342 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 12 |
Hb_001269_380 |
0.0732031053 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 13 |
Hb_019280_010 |
0.0732308971 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003544_050 |
0.0733451307 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006452_120 |
0.0743207922 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 16 |
Hb_004030_080 |
0.0745325486 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 17 |
Hb_000358_050 |
0.0752986851 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 18 |
Hb_008948_020 |
0.0761024111 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 19 |
Hb_002908_050 |
0.0763602589 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 20 |
Hb_000363_040 |
0.0766603392 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |