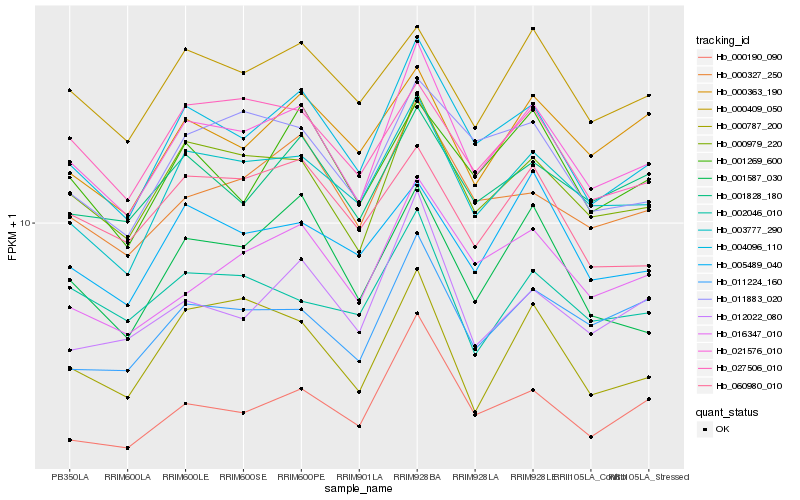
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021576_010 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 2 |
Hb_000979_220 |
0.0449438333 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004096_110 |
0.0452133677 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 4 |
Hb_003777_290 |
0.0554660457 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 5 |
Hb_000327_250 |
0.0690037984 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 6 |
Hb_016347_010 |
0.0738009337 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 7 |
Hb_001828_180 |
0.0768744702 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 8 |
Hb_011224_160 |
0.077383593 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 9 |
Hb_011883_020 |
0.0785595904 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 10 |
Hb_000409_050 |
0.0792789373 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 11 |
Hb_000190_090 |
0.0802982846 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 12 |
Hb_001587_030 |
0.0817944098 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 13 |
Hb_027506_010 |
0.0818647586 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 14 |
Hb_012022_080 |
0.0828805426 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_001269_600 |
0.0844806293 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 16 |
Hb_002046_010 |
0.0848339814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_060980_010 |
0.0855189002 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000363_190 |
0.0858582209 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005489_040 |
0.0860268864 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 20 |
Hb_000787_200 |
0.0861345089 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |