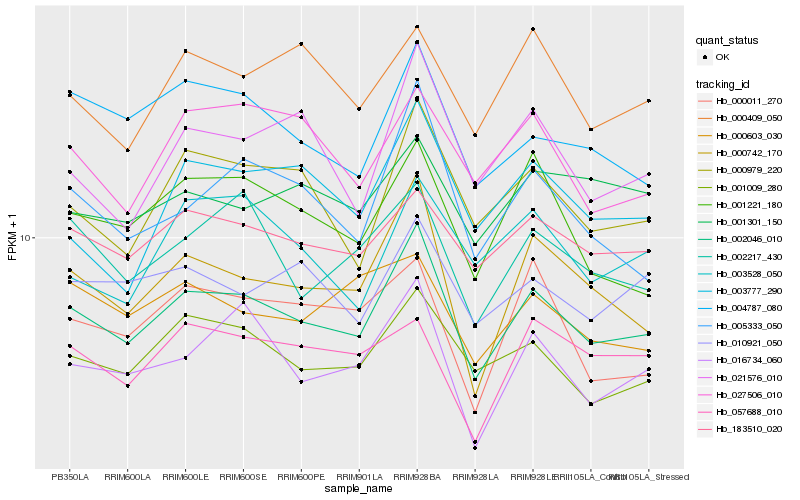
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002046_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000742_170 |
0.065714732 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_001009_280 |
0.0698268203 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 4 |
Hb_000979_220 |
0.0738693843 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 5 |
Hb_183510_020 |
0.0754138649 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 6 |
Hb_016734_060 |
0.0761191801 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 7 |
Hb_005333_050 |
0.0774268377 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004787_080 |
0.0794641419 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 9 |
Hb_001221_180 |
0.0810222287 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 10 |
Hb_002217_430 |
0.0826745254 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 11 |
Hb_021576_010 |
0.0848339814 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 12 |
Hb_010921_050 |
0.0848990943 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003777_290 |
0.0873527719 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 14 |
Hb_027506_010 |
0.0879624219 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 15 |
Hb_000409_050 |
0.090303535 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 16 |
Hb_057688_010 |
0.0922455529 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001301_150 |
0.093172975 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 18 |
Hb_003528_050 |
0.0938459188 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 19 |
Hb_000011_270 |
0.0939466417 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 20 |
Hb_000603_030 |
0.0939572896 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |