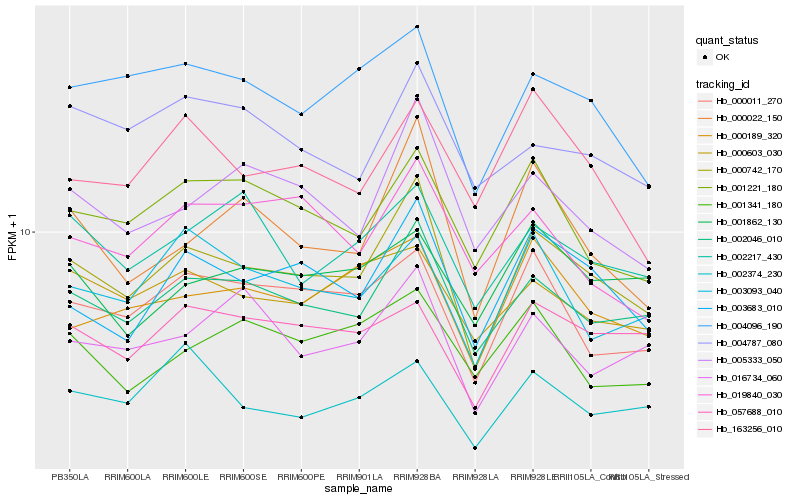
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_170 |
0.0 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_002046_010 |
0.065714732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003683_010 |
0.0786717647 |
- |
- |
longevity assurance factor, putative [Ricinus communis] |
| 4 |
Hb_000022_150 |
0.0908734344 |
- |
- |
- |
| 5 |
Hb_005333_050 |
0.0922235315 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000011_270 |
0.0968420419 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 7 |
Hb_004096_190 |
0.0968455492 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 8 |
Hb_001221_180 |
0.0997112189 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_000189_320 |
0.1016120801 |
- |
- |
PREDICTED: multisubstrate pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 10 |
Hb_016734_060 |
0.1054732709 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 11 |
Hb_057688_010 |
0.107917155 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003093_040 |
0.1082292484 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_002217_430 |
0.1103756669 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 14 |
Hb_000603_030 |
0.1112100662 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 15 |
Hb_163256_010 |
0.1124307591 |
- |
- |
PREDICTED: phosphoglucomutase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004787_080 |
0.1134213013 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 17 |
Hb_019840_030 |
0.1145244888 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001862_130 |
0.1163500906 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 19 |
Hb_002374_230 |
0.1171941411 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 20 |
Hb_001341_180 |
0.1172536812 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |