| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
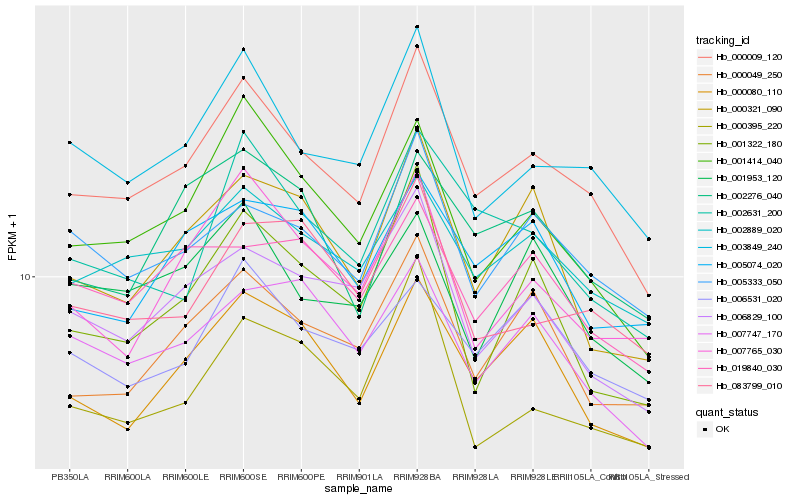
Hb_000009_120 |
0.0 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 2 |
Hb_003849_240 |
0.0687556165 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_001414_040 |
0.0817664262 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002889_020 |
0.0819363132 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 5 |
Hb_005333_050 |
0.0875794567 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_006829_100 |
0.0883018728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000080_110 |
0.0909830906 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000049_250 |
0.0919639305 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002631_200 |
0.0969851923 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007765_030 |
0.0993097078 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 11 |
Hb_000321_090 |
0.1007601618 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 12 |
Hb_000395_220 |
0.1011471561 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 13 |
Hb_007747_170 |
0.1025206226 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 14 |
Hb_001322_180 |
0.1025265953 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 15 |
Hb_002276_040 |
0.1063604778 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 16 |
Hb_001953_120 |
0.1065306283 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006531_020 |
0.1066920835 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 18 |
Hb_083799_010 |
0.1068500257 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_005074_020 |
0.1076495543 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 20 |
Hb_019840_030 |
0.1082582907 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |