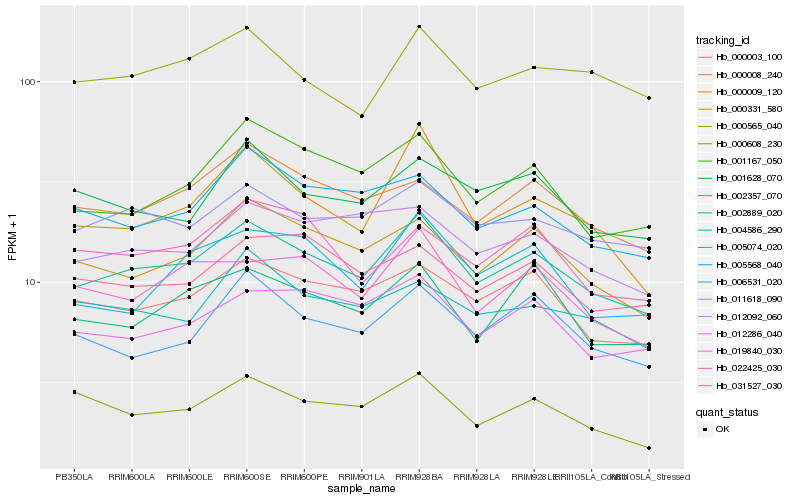
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002889_020 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 2 |
Hb_000003_100 |
0.0695975904 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 3 |
Hb_000331_580 |
0.0697865817 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 4 |
Hb_011618_090 |
0.0701869344 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_012286_040 |
0.0756009252 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 6 |
Hb_019840_030 |
0.0760193866 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001167_050 |
0.0770008083 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 8 |
Hb_000008_240 |
0.0776805589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006531_020 |
0.0780129607 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 10 |
Hb_000565_040 |
0.0793059085 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 11 |
Hb_002357_070 |
0.0802109621 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 12 |
Hb_012092_060 |
0.0804760898 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 13 |
Hb_005568_040 |
0.0814072414 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000009_120 |
0.0819363132 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 15 |
Hb_001628_070 |
0.0828471247 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_031527_030 |
0.0835267828 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 17 |
Hb_005074_020 |
0.0843253965 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000608_230 |
0.0849939951 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 19 |
Hb_004586_290 |
0.0855343138 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 20 |
Hb_022425_030 |
0.0862445411 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |