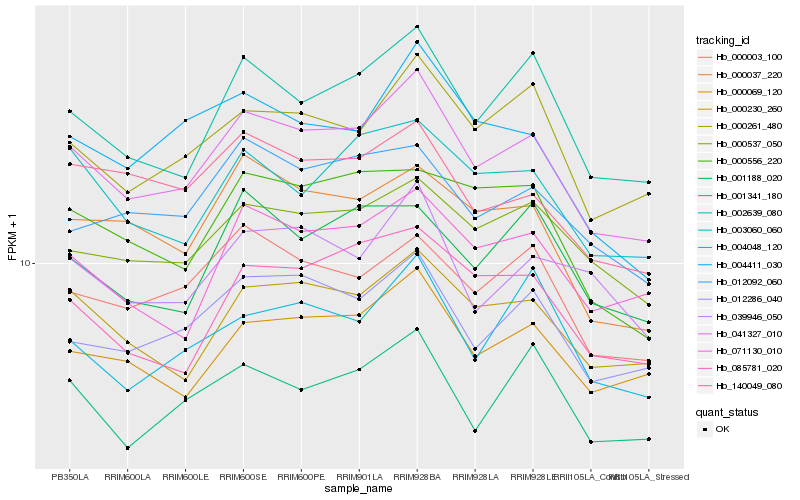
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041327_010 |
0.0 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 2 |
Hb_002639_080 |
0.0550412216 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012286_040 |
0.0578915572 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 4 |
Hb_000261_480 |
0.0582591124 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 5 |
Hb_000230_260 |
0.0590361267 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 6 |
Hb_000069_120 |
0.060001994 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 7 |
Hb_000537_050 |
0.0654627145 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 8 |
Hb_000003_100 |
0.0658301423 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 9 |
Hb_071130_010 |
0.0662128287 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003060_060 |
0.0683026462 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 11 |
Hb_000037_220 |
0.0701289807 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 12 |
Hb_085781_020 |
0.0704668755 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 13 |
Hb_001341_180 |
0.0715391139 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 14 |
Hb_140049_080 |
0.0717346154 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 15 |
Hb_012092_060 |
0.0731350914 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 16 |
Hb_039946_050 |
0.0735589172 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_004411_030 |
0.0750005963 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 18 |
Hb_004048_120 |
0.0755273171 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 19 |
Hb_001188_020 |
0.0761724983 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000556_220 |
0.0763191435 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |