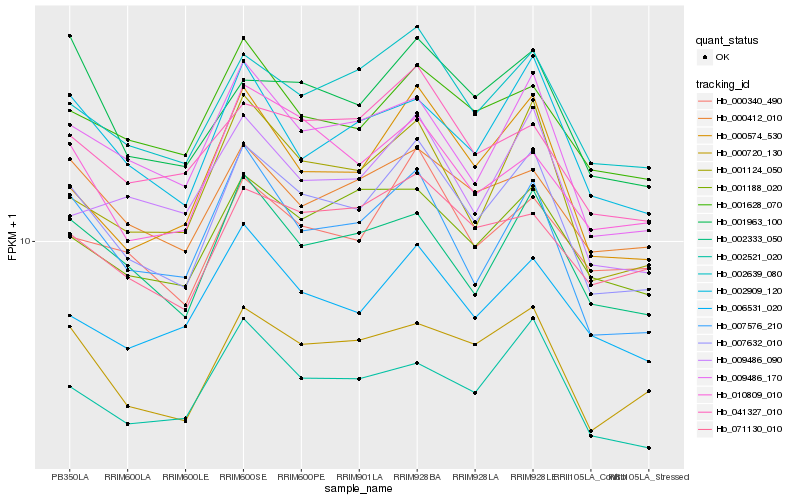
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007632_010 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002639_080 |
0.0656161673 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000720_130 |
0.0692315021 |
- |
- |
hypothetical protein PRUPE_ppa012392mg [Prunus persica] |
| 4 |
Hb_000574_530 |
0.0707016349 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 5 |
Hb_007576_210 |
0.0708788087 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 6 |
Hb_009486_170 |
0.0714203151 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 7 |
Hb_002333_050 |
0.0754118081 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001124_050 |
0.0755110148 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 9 |
Hb_002909_120 |
0.0783969138 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 10 |
Hb_000340_490 |
0.0821902125 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 11 |
Hb_002521_020 |
0.0825760896 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001963_100 |
0.083473612 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 13 |
Hb_001628_070 |
0.0837866868 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_000412_010 |
0.0838694418 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_001188_020 |
0.0844432484 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006531_020 |
0.0847932403 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 17 |
Hb_041327_010 |
0.0870564735 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 18 |
Hb_009486_090 |
0.0881125963 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_071130_010 |
0.0885090883 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010809_010 |
0.0891173938 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |