| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
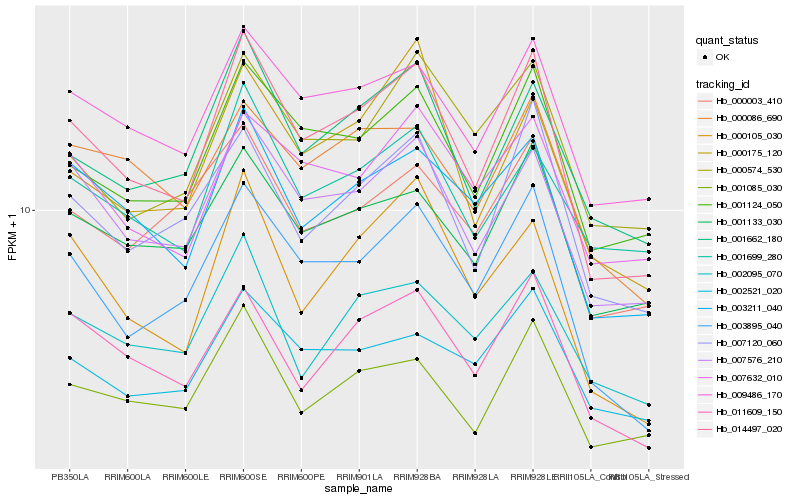
Hb_014497_020 |
0.0 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 2 |
Hb_007576_210 |
0.0654435147 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 3 |
Hb_003895_040 |
0.0669059705 |
- |
- |
hypothetical protein CISIN_1g0006512mg, partial [Citrus sinensis] |
| 4 |
Hb_001124_050 |
0.0712677421 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 5 |
Hb_003211_040 |
0.0778107441 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001662_180 |
0.0783142158 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 7 |
Hb_009486_170 |
0.0797537291 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 8 |
Hb_001085_030 |
0.0811347662 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_007120_060 |
0.0849095747 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 10 |
Hb_001133_030 |
0.0861366663 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 11 |
Hb_011609_150 |
0.0872950952 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |
| 12 |
Hb_000105_030 |
0.0904127554 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 13 |
Hb_000175_120 |
0.0908122314 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 14 |
Hb_002521_020 |
0.0941294895 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_007632_010 |
0.0969175947 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000086_690 |
0.0969279343 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 17 |
Hb_002095_070 |
0.0979987522 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 18 |
Hb_001699_280 |
0.0983535477 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 19 |
Hb_000574_530 |
0.098646176 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 20 |
Hb_000003_410 |
0.0988514989 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |