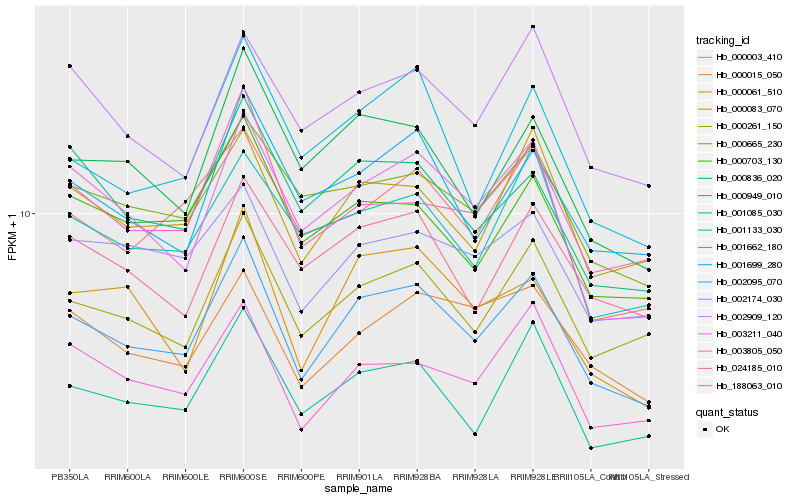
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002095_070 |
0.0 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 2 |
Hb_000261_150 |
0.0726735433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003211_040 |
0.0727350223 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000083_070 |
0.0791822598 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000836_020 |
0.0827162944 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 6 |
Hb_003805_050 |
0.0831528315 |
- |
- |
- |
| 7 |
Hb_001085_030 |
0.0842563393 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 8 |
Hb_001699_280 |
0.0851080889 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 9 |
Hb_001662_180 |
0.0852547859 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 10 |
Hb_000061_510 |
0.0861307042 |
- |
- |
hypothetical protein JCGZ_11686 [Jatropha curcas] |
| 11 |
Hb_000015_050 |
0.0862037078 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 12 |
Hb_000703_130 |
0.08696426 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 13 |
Hb_188063_010 |
0.0873590055 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 14 |
Hb_000949_010 |
0.0875262507 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 15 |
Hb_001133_030 |
0.0876435326 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 16 |
Hb_000665_230 |
0.0878142004 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 17 |
Hb_002909_120 |
0.0879240323 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 18 |
Hb_002174_030 |
0.0879600775 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 19 |
Hb_000003_410 |
0.0888588444 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 20 |
Hb_024185_010 |
0.0888682677 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |