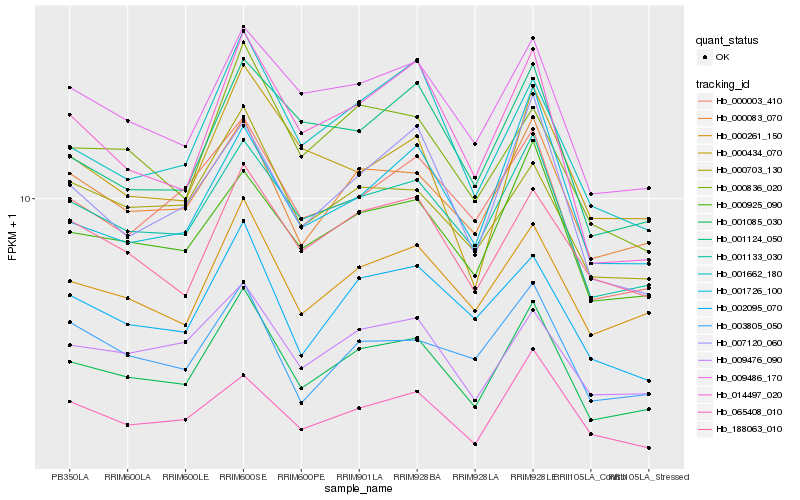
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_030 |
0.0 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_001133_030 |
0.0710606977 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 3 |
Hb_000261_150 |
0.0775950218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000925_090 |
0.0791626668 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000083_070 |
0.0792058756 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007120_060 |
0.0800240526 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 7 |
Hb_014497_020 |
0.0811347662 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 8 |
Hb_009476_090 |
0.0821692543 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 9 |
Hb_002095_070 |
0.0842563393 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 10 |
Hb_001662_180 |
0.0869490526 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 11 |
Hb_009486_170 |
0.0886269599 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 12 |
Hb_000003_410 |
0.0898788705 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 13 |
Hb_065408_010 |
0.0911807441 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000836_020 |
0.0920154533 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 15 |
Hb_000703_130 |
0.093144256 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 16 |
Hb_001726_100 |
0.0939215822 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001124_050 |
0.0942598422 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 18 |
Hb_000434_070 |
0.0960505444 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 19 |
Hb_003805_050 |
0.096118279 |
- |
- |
- |
| 20 |
Hb_188063_010 |
0.0975333518 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |