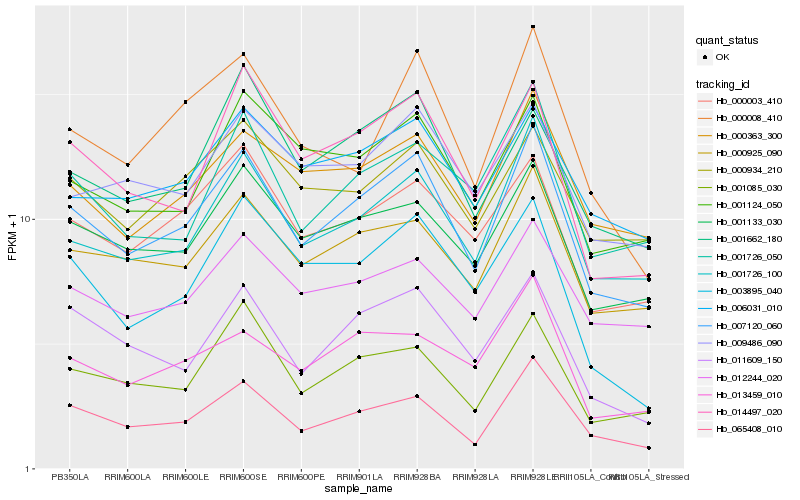
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007120_060 |
0.0 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 2 |
Hb_065408_010 |
0.0634215934 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001726_100 |
0.067578617 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000925_090 |
0.0720552576 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001133_030 |
0.0761188616 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 6 |
Hb_001085_030 |
0.0800240526 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 7 |
Hb_000003_410 |
0.0820307779 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 8 |
Hb_000363_300 |
0.0834898693 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011609_150 |
0.0847619372 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |
| 10 |
Hb_014497_020 |
0.0849095747 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 11 |
Hb_001124_050 |
0.0895927175 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 12 |
Hb_001726_050 |
0.0907654102 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 13 |
Hb_012244_020 |
0.0910488694 |
- |
- |
calpain, putative [Ricinus communis] |
| 14 |
Hb_013459_010 |
0.0917148046 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 15 |
Hb_006031_010 |
0.0919031727 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 16 |
Hb_009486_090 |
0.0924735328 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_000934_210 |
0.0941193665 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 18 |
Hb_001662_180 |
0.0952614328 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 19 |
Hb_000008_410 |
0.0963098556 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_003895_040 |
0.096871019 |
- |
- |
hypothetical protein CISIN_1g0006512mg, partial [Citrus sinensis] |