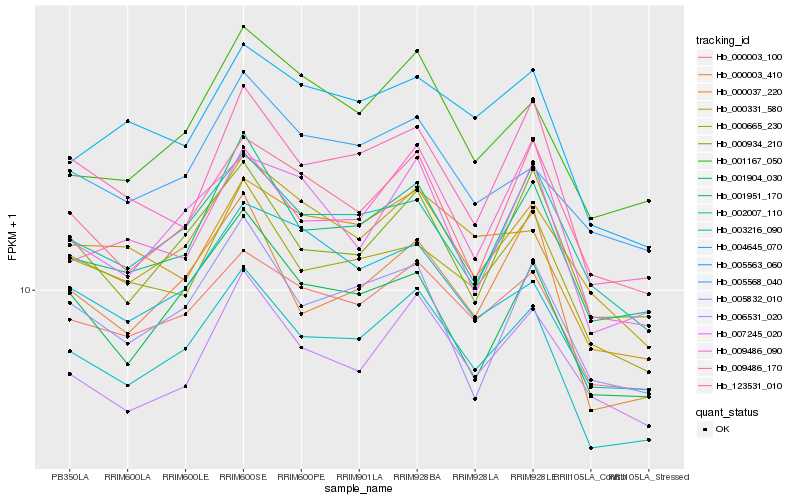
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003216_090 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000003_100 |
0.0495378889 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 3 |
Hb_001951_170 |
0.051403809 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 4 |
Hb_000331_580 |
0.0585964133 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 5 |
Hb_000003_410 |
0.0647838944 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 6 |
Hb_000665_230 |
0.0670549006 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 7 |
Hb_001904_030 |
0.0678928639 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 8 |
Hb_000934_210 |
0.0686714789 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 9 |
Hb_009486_170 |
0.0693114915 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 10 |
Hb_001167_050 |
0.0694225047 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 11 |
Hb_002007_110 |
0.0705088419 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005568_040 |
0.0707367978 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005832_010 |
0.0712774511 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004645_070 |
0.0718354842 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 15 |
Hb_006531_020 |
0.0719089705 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 16 |
Hb_005563_060 |
0.072259484 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 17 |
Hb_007245_020 |
0.0722964368 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 18 |
Hb_123531_010 |
0.0729785227 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 19 |
Hb_009486_090 |
0.0742071088 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000037_220 |
0.0743130702 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |