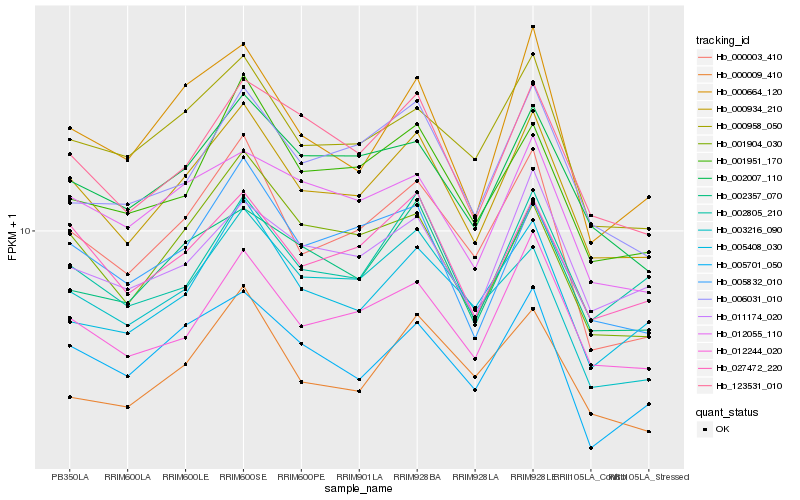
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000934_210 |
0.0 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 2 |
Hb_027472_220 |
0.0504158595 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_012244_020 |
0.0525087545 |
- |
- |
calpain, putative [Ricinus communis] |
| 4 |
Hb_123531_010 |
0.0534549689 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 5 |
Hb_002007_110 |
0.0556618555 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002357_070 |
0.0569379347 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 7 |
Hb_000958_050 |
0.0590682127 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 8 |
Hb_005832_010 |
0.0616218268 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001951_170 |
0.0620135896 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 10 |
Hb_000003_410 |
0.0633501927 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 11 |
Hb_005701_050 |
0.064072391 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 12 |
Hb_011174_020 |
0.0640792578 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 13 |
Hb_012055_110 |
0.0640902791 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 14 |
Hb_006031_010 |
0.0647637613 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 15 |
Hb_001904_030 |
0.0663938638 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 16 |
Hb_002805_210 |
0.0668382226 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 17 |
Hb_005408_030 |
0.0674932241 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 18 |
Hb_000009_410 |
0.0683640872 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 19 |
Hb_003216_090 |
0.0686714789 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000664_120 |
0.069028233 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |