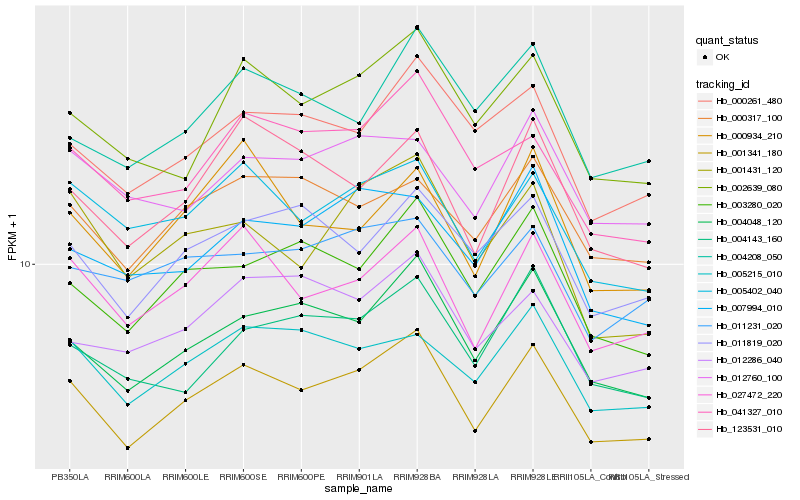
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001341_180 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 2 |
Hb_004048_120 |
0.0549572672 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 3 |
Hb_027472_220 |
0.0589304675 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_003280_020 |
0.0668074726 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005402_040 |
0.0679856406 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 6 |
Hb_002639_080 |
0.0681097119 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000261_480 |
0.0697498057 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 8 |
Hb_041327_010 |
0.0715391139 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 9 |
Hb_005215_010 |
0.071702213 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011819_020 |
0.0724240502 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000317_100 |
0.0733674758 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 12 |
Hb_123531_010 |
0.074144266 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 13 |
Hb_004143_160 |
0.0752495313 |
- |
- |
SAB, putative [Ricinus communis] |
| 14 |
Hb_007994_010 |
0.0772415703 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_012760_100 |
0.0777006626 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 16 |
Hb_004208_050 |
0.0778123476 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 17 |
Hb_012286_040 |
0.0780701647 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 18 |
Hb_001431_120 |
0.0788135339 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 19 |
Hb_011231_020 |
0.0794905808 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000934_210 |
0.0807160967 |
- |
- |
suppressor of ty, putative [Ricinus communis] |