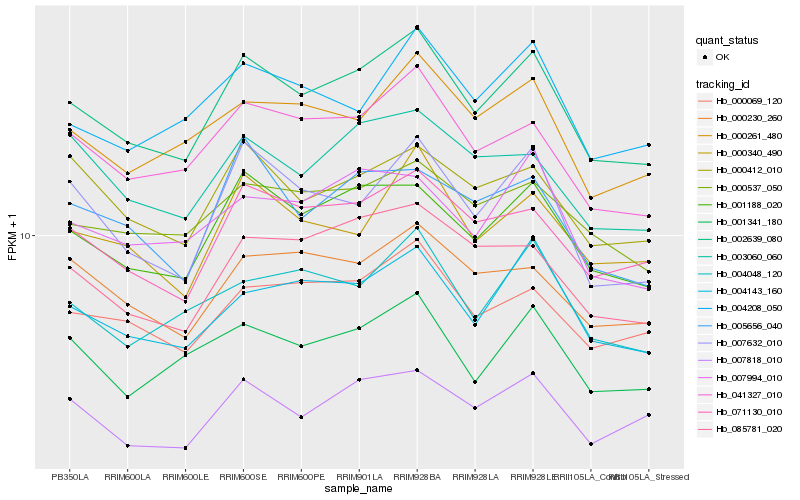
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_080 |
0.0 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001188_020 |
0.0479061481 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_041327_010 |
0.0550412216 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 4 |
Hb_071130_010 |
0.0563175483 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004143_160 |
0.0642042086 |
- |
- |
SAB, putative [Ricinus communis] |
| 6 |
Hb_000069_120 |
0.064771933 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 7 |
Hb_007632_010 |
0.0656161673 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000261_480 |
0.0660441977 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 9 |
Hb_000340_490 |
0.0661587112 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 10 |
Hb_085781_020 |
0.0680533235 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 11 |
Hb_001341_180 |
0.0681097119 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 12 |
Hb_000537_050 |
0.0704670632 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 13 |
Hb_007818_010 |
0.0706765669 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like isoform X2 [Gossypium raimondii] |
| 14 |
Hb_004208_050 |
0.0717163676 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 15 |
Hb_004048_120 |
0.072196342 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000412_010 |
0.0725496165 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_000230_260 |
0.0734845156 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 18 |
Hb_003060_060 |
0.0742435746 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 19 |
Hb_007994_010 |
0.0742832729 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005656_040 |
0.075758614 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |