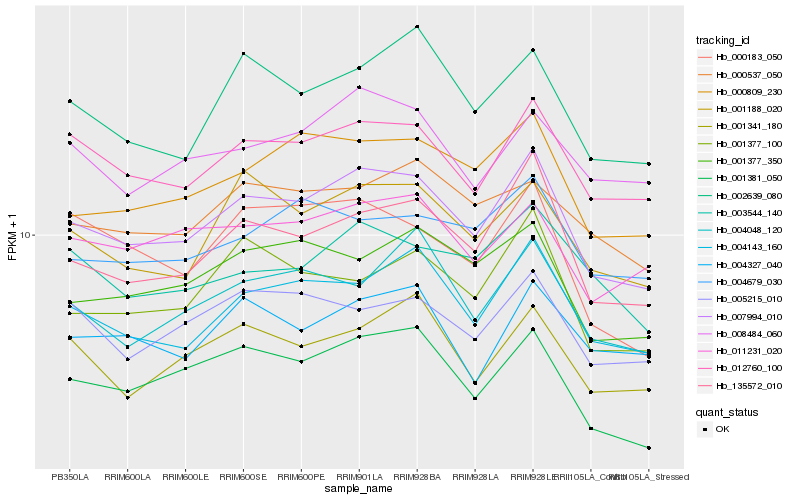
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007994_010 |
0.0 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_135572_010 |
0.0522170325 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 3 |
Hb_012760_100 |
0.0586281599 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 4 |
Hb_004143_160 |
0.0601611014 |
- |
- |
SAB, putative [Ricinus communis] |
| 5 |
Hb_001381_050 |
0.0650914893 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_000809_230 |
0.0659594158 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001188_020 |
0.0686077572 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001377_350 |
0.0731119576 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 9 |
Hb_002639_080 |
0.0742832729 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005215_010 |
0.0763778576 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_011231_020 |
0.0767381973 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001377_100 |
0.076994151 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001341_180 |
0.0772415703 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 14 |
Hb_008484_060 |
0.0776954059 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 15 |
Hb_004048_120 |
0.0778873725 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 16 |
Hb_004679_030 |
0.0795640117 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 17 |
Hb_003544_140 |
0.0806542164 |
- |
- |
PREDICTED: protein GDAP2 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_000537_050 |
0.0813016583 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 19 |
Hb_000183_050 |
0.0816362351 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 20 |
Hb_004327_040 |
0.0835697568 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |