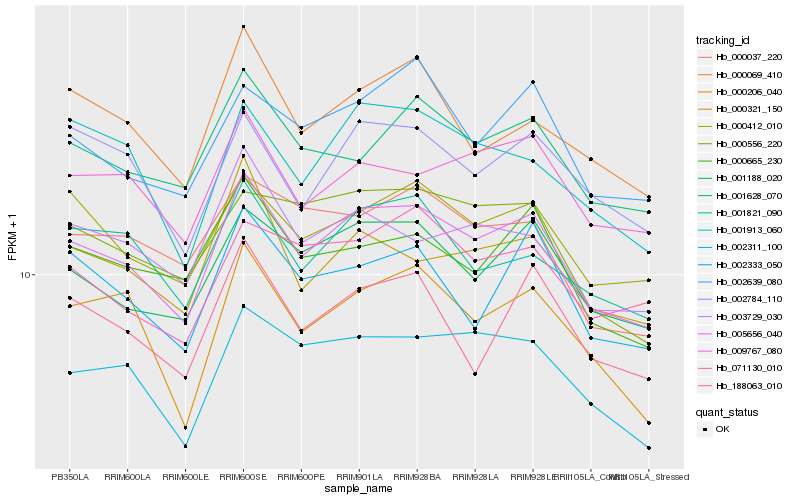
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005656_040 |
0.0 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 2 |
Hb_001188_020 |
0.0598506734 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000412_010 |
0.0660561403 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_002311_100 |
0.0669826481 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 5 |
Hb_001628_070 |
0.0728426173 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_009767_080 |
0.0728777702 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 7 |
Hb_000206_040 |
0.0747764717 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 8 |
Hb_000665_230 |
0.074825243 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 9 |
Hb_001821_090 |
0.0751877091 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 10 |
Hb_002639_080 |
0.075758614 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000069_410 |
0.0768665622 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 12 |
Hb_000556_220 |
0.0769540153 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 13 |
Hb_000321_150 |
0.0769622841 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003729_030 |
0.0774632954 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 15 |
Hb_002784_110 |
0.0776237094 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 16 |
Hb_002333_050 |
0.0792728314 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 17 |
Hb_188063_010 |
0.0794234565 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 18 |
Hb_001913_060 |
0.0795798402 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 19 |
Hb_000037_220 |
0.0801785172 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 20 |
Hb_071130_010 |
0.0802186828 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |