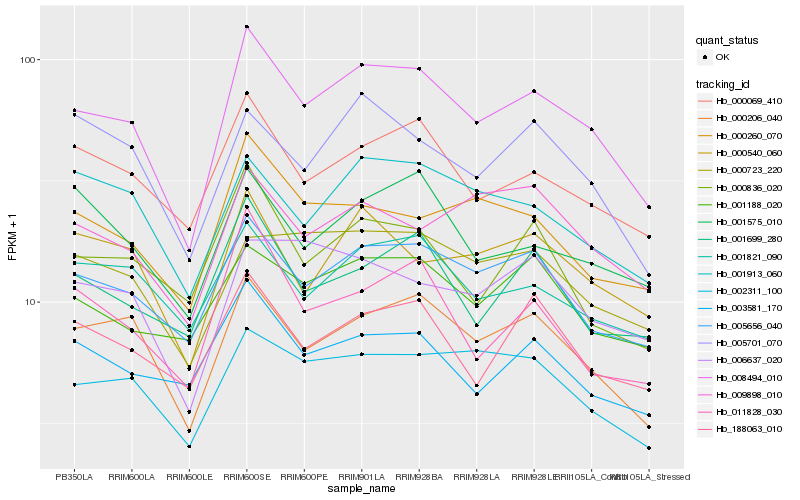
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008494_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 2 |
Hb_000206_040 |
0.0718049166 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 3 |
Hb_005656_040 |
0.0804826898 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_002311_100 |
0.0909887264 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 5 |
Hb_000069_410 |
0.0911733648 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 6 |
Hb_000540_060 |
0.0986807668 |
- |
- |
PREDICTED: uncharacterized protein LOC105628177 [Jatropha curcas] |
| 7 |
Hb_001913_060 |
0.1011908788 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 8 |
Hb_000260_070 |
0.1018514237 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_001821_090 |
0.1027039399 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 10 |
Hb_188063_010 |
0.1033452752 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 11 |
Hb_001699_280 |
0.1036827823 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 12 |
Hb_000723_220 |
0.103967303 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 13 |
Hb_001188_020 |
0.1052874459 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001575_010 |
0.105845392 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_011828_030 |
0.1060939697 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 16 |
Hb_000836_020 |
0.1063897711 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 17 |
Hb_005701_070 |
0.1064076352 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 18 |
Hb_003581_170 |
0.1069668287 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006637_020 |
0.1074769381 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 20 |
Hb_009898_010 |
0.1075709367 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |