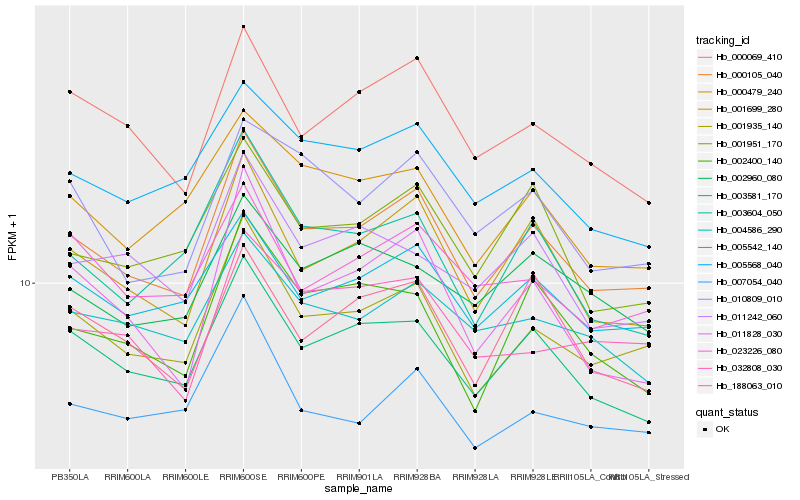
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000105_040 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 2 |
Hb_001935_140 |
0.0412483516 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 3 |
Hb_001699_280 |
0.0476890025 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 4 |
Hb_003581_170 |
0.0523856452 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005542_140 |
0.0666634702 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 6 |
Hb_000069_410 |
0.0697591613 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 7 |
Hb_001951_170 |
0.0709622939 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 8 |
Hb_005568_040 |
0.0709839109 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000479_240 |
0.0737164354 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002400_140 |
0.0739321205 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 11 |
Hb_003604_050 |
0.0744769442 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_004586_290 |
0.076844559 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 13 |
Hb_188063_010 |
0.0772728487 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 14 |
Hb_011828_030 |
0.0775090133 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 15 |
Hb_010809_010 |
0.0776258616 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |
| 16 |
Hb_007054_040 |
0.0777609786 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 17 |
Hb_011242_060 |
0.080237053 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 18 |
Hb_023226_080 |
0.0803794688 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002960_080 |
0.0817246847 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 20 |
Hb_032808_030 |
0.0817540599 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |