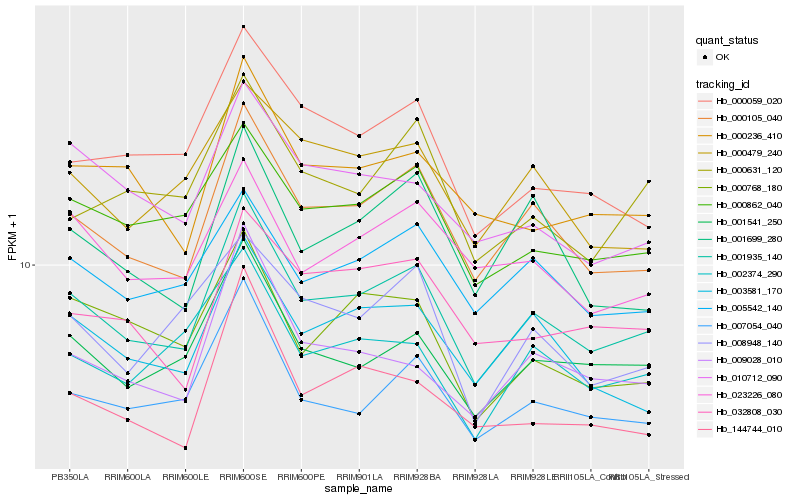
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001935_140 |
0.0 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 2 |
Hb_000105_040 |
0.0412483516 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 3 |
Hb_001541_250 |
0.0633552127 |
- |
- |
PREDICTED: integrator complex subunit 9 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_007054_040 |
0.0655680817 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 5 |
Hb_000862_040 |
0.0693800791 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 6 |
Hb_003581_170 |
0.07143426 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005542_140 |
0.0726375453 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 8 |
Hb_009028_010 |
0.0767539799 |
- |
- |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 9 |
Hb_000236_410 |
0.0798590051 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 10 |
Hb_001699_280 |
0.0803567352 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 11 |
Hb_000479_240 |
0.0803896373 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_144744_010 |
0.0808544722 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 13 |
Hb_000059_020 |
0.0816415666 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 14 |
Hb_002374_290 |
0.0821409697 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 15 |
Hb_032808_030 |
0.0825863671 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_023226_080 |
0.082796511 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000631_120 |
0.0833839053 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 18 |
Hb_010712_090 |
0.0835077819 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 19 |
Hb_000768_180 |
0.0842441111 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 20 |
Hb_008948_140 |
0.0857916068 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |