| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
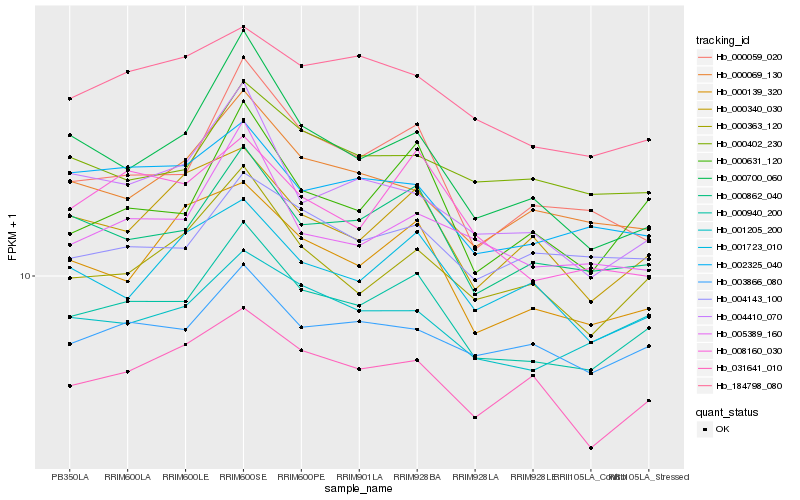
Hb_000940_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 2 |
Hb_000862_040 |
0.0536061139 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 3 |
Hb_000631_120 |
0.0616600247 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 4 |
Hb_001205_200 |
0.068395779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000363_120 |
0.0691968068 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 6 |
Hb_000059_020 |
0.0708684763 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 7 |
Hb_003866_080 |
0.0739845315 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 8 |
Hb_031641_010 |
0.0759941895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000069_130 |
0.076136726 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005389_160 |
0.0763861926 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 11 |
Hb_002325_040 |
0.0769396253 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000139_320 |
0.0771791807 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 13 |
Hb_184798_080 |
0.0772059574 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 14 |
Hb_000340_030 |
0.0796859603 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 15 |
Hb_008160_030 |
0.0802829982 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_004410_070 |
0.0808337439 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 17 |
Hb_000402_230 |
0.0813193606 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001723_010 |
0.0819063025 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 19 |
Hb_004143_100 |
0.0819080151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000700_060 |
0.0831511581 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |