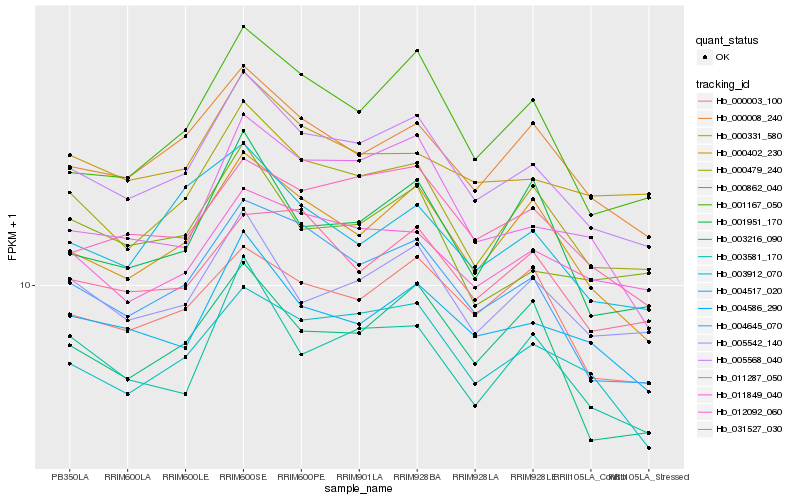
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005568_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000479_240 |
0.039694625 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004645_070 |
0.0512381923 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 4 |
Hb_000331_580 |
0.0516715073 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 5 |
Hb_000008_240 |
0.0536660326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003912_070 |
0.0564260511 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 7 |
Hb_004586_290 |
0.0565737843 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 8 |
Hb_005542_140 |
0.0597516877 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 9 |
Hb_012092_060 |
0.0612462583 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 10 |
Hb_001167_050 |
0.0619087238 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 11 |
Hb_000003_100 |
0.0651118038 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 12 |
Hb_011849_040 |
0.0666597357 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_003581_170 |
0.0667526962 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_011287_050 |
0.0682496991 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 15 |
Hb_031527_030 |
0.0692192652 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 16 |
Hb_000402_230 |
0.0693228654 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004517_020 |
0.0693378776 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 18 |
Hb_000862_040 |
0.069884863 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 19 |
Hb_001951_170 |
0.0704741539 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 20 |
Hb_003216_090 |
0.0707367978 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |