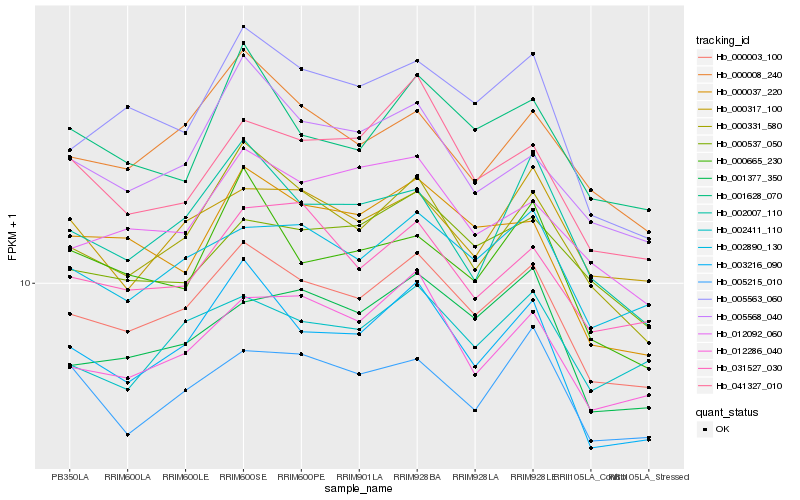
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_100 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 2 |
Hb_003216_090 |
0.0495378889 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000331_580 |
0.0503594872 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 4 |
Hb_002890_130 |
0.0536226944 |
- |
- |
tip120, putative [Ricinus communis] |
| 5 |
Hb_005563_060 |
0.0569663576 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 6 |
Hb_012286_040 |
0.0580103665 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 7 |
Hb_000008_240 |
0.0591953989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000037_220 |
0.0604281996 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 9 |
Hb_031527_030 |
0.0617177746 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 10 |
Hb_001377_350 |
0.0618323466 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 11 |
Hb_001628_070 |
0.0626512968 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_012092_060 |
0.0636427728 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 13 |
Hb_002007_110 |
0.0644082453 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 14 |
Hb_005568_040 |
0.0651118038 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 15 |
Hb_041327_010 |
0.0658301423 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 16 |
Hb_000537_050 |
0.0662632147 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 17 |
Hb_000317_100 |
0.0663326434 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 18 |
Hb_000665_230 |
0.0669452788 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 19 |
Hb_002411_110 |
0.0673674292 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 20 |
Hb_005215_010 |
0.0676700911 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |