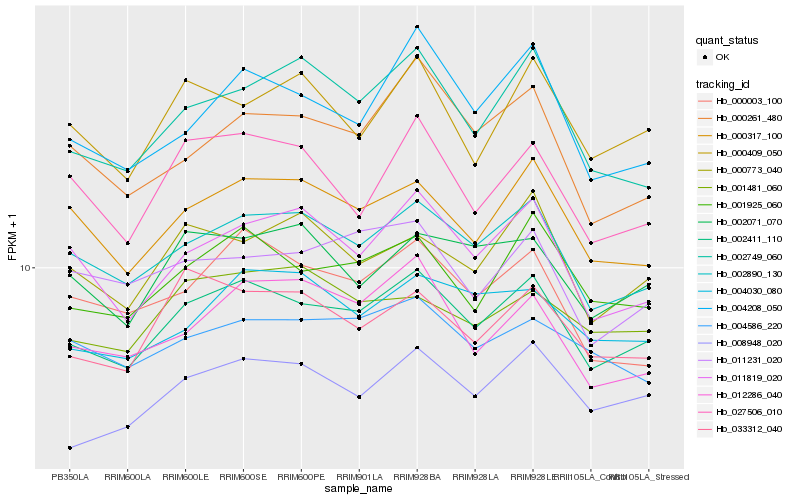
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002411_110 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 2 |
Hb_002890_130 |
0.0420732874 |
- |
- |
tip120, putative [Ricinus communis] |
| 3 |
Hb_001925_060 |
0.0486791108 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 4 |
Hb_008948_020 |
0.0560331809 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 5 |
Hb_000317_100 |
0.0563933768 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 6 |
Hb_011819_020 |
0.0607865215 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 7 |
Hb_033312_040 |
0.062258088 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004208_050 |
0.0641702325 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 9 |
Hb_002071_070 |
0.0663904074 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004030_080 |
0.0664243025 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 11 |
Hb_002749_060 |
0.0667995669 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 12 |
Hb_000773_040 |
0.066870207 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 13 |
Hb_012286_040 |
0.0669731792 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 14 |
Hb_027506_010 |
0.067149656 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 15 |
Hb_004586_220 |
0.0672786906 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000003_100 |
0.0673674292 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 17 |
Hb_000261_480 |
0.0678636485 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 18 |
Hb_011231_020 |
0.0689577767 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001481_060 |
0.0692370834 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000409_050 |
0.0698922631 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |