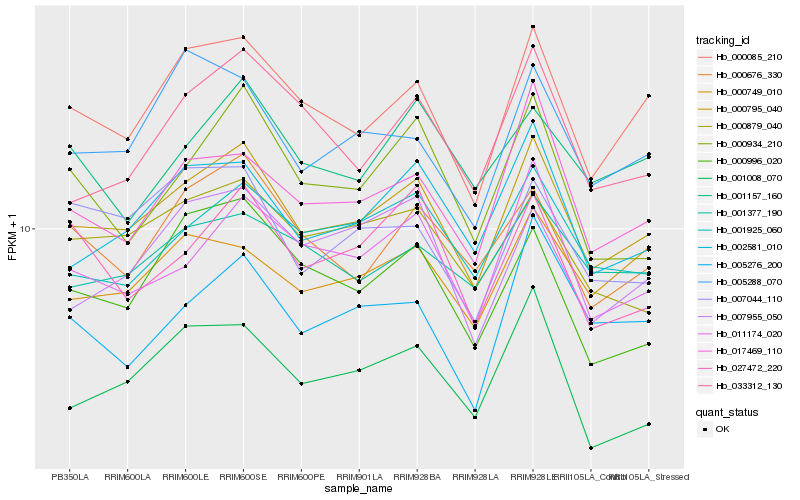
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000795_040 |
0.0 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 2 |
Hb_000085_210 |
0.0503472261 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 3 |
Hb_002581_010 |
0.0571986333 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 4 |
Hb_011174_020 |
0.060133997 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 5 |
Hb_017469_110 |
0.0607234657 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 6 |
Hb_001925_060 |
0.0630811266 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 7 |
Hb_000879_040 |
0.0660874549 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 8 |
Hb_001008_070 |
0.0664222352 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 9 |
Hb_000676_330 |
0.0676559883 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_001377_190 |
0.0685247454 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005288_070 |
0.0698054694 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000749_010 |
0.0703225903 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_033312_130 |
0.0715405188 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 14 |
Hb_007955_050 |
0.072638113 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_000996_020 |
0.0726626891 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 16 |
Hb_005276_200 |
0.0734538685 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000934_210 |
0.0752971885 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 18 |
Hb_001157_160 |
0.0759036432 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007044_110 |
0.0760270878 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_027472_220 |
0.0761053553 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |