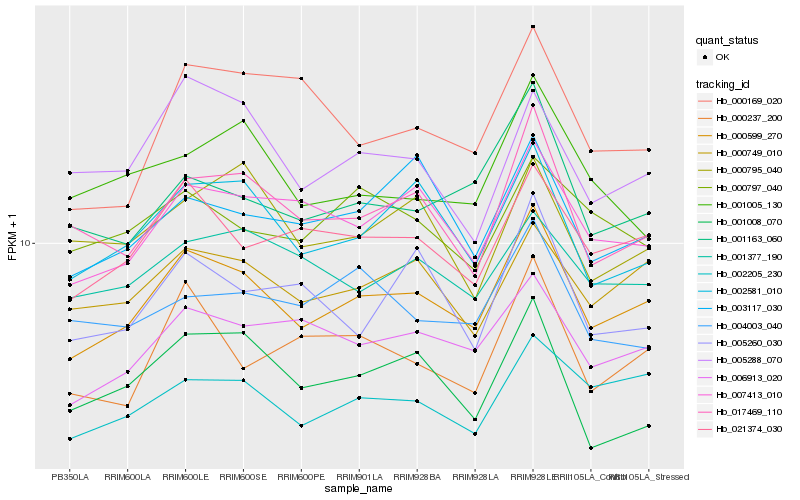
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_270 |
0.0 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 2 |
Hb_021374_030 |
0.0640696186 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 3 |
Hb_002581_010 |
0.0653828074 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006913_020 |
0.0660200102 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007413_010 |
0.0682686686 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 6 |
Hb_017469_110 |
0.0683763698 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 7 |
Hb_002205_230 |
0.0688196503 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000749_010 |
0.0689493441 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001163_060 |
0.0756997656 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 10 |
Hb_003117_030 |
0.0796389027 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 11 |
Hb_001377_190 |
0.081063753 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000237_200 |
0.0833972388 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 13 |
Hb_005288_070 |
0.0842516405 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_001008_070 |
0.0847023532 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 15 |
Hb_000797_040 |
0.087038595 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 16 |
Hb_005260_030 |
0.0874392624 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 17 |
Hb_000795_040 |
0.0891203494 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 18 |
Hb_004003_040 |
0.090349529 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_000169_020 |
0.0904241962 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 20 |
Hb_001005_130 |
0.0905131732 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |