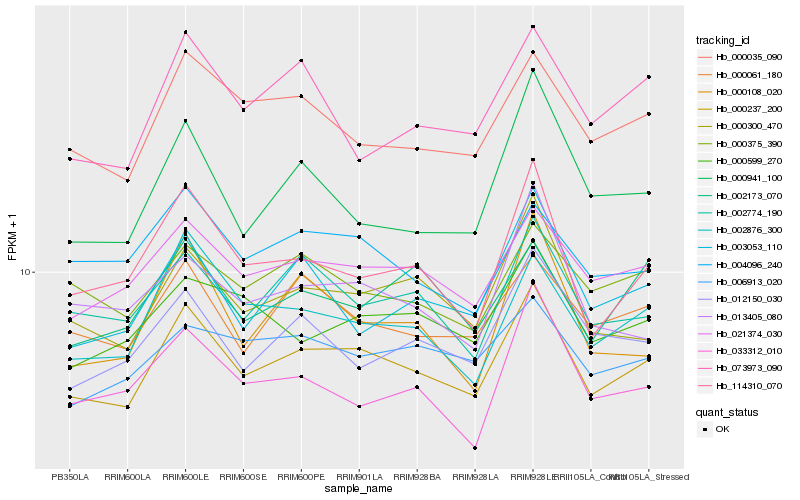
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_200 |
0.0 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 2 |
Hb_021374_030 |
0.0667435258 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 3 |
Hb_000035_090 |
0.0731214717 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 4 |
Hb_000300_470 |
0.0745376323 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 5 |
Hb_002876_300 |
0.075061918 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_114310_070 |
0.075933774 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 7 |
Hb_002774_190 |
0.0766852527 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 8 |
Hb_000061_180 |
0.0776349468 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 9 |
Hb_000599_270 |
0.0833972388 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 10 |
Hb_012150_030 |
0.0859397593 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003053_110 |
0.0862432035 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000375_390 |
0.0869743488 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 13 |
Hb_004096_240 |
0.0883202649 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 14 |
Hb_006913_020 |
0.0891380144 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 15 |
Hb_073973_090 |
0.0903827838 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 16 |
Hb_000941_100 |
0.0903973258 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 17 |
Hb_013405_080 |
0.0916058909 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_000108_020 |
0.0919094399 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 19 |
Hb_002173_070 |
0.0921168467 |
- |
- |
MRG family protein isoform 2 [Theobroma cacao] |
| 20 |
Hb_033312_010 |
0.0924053148 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |