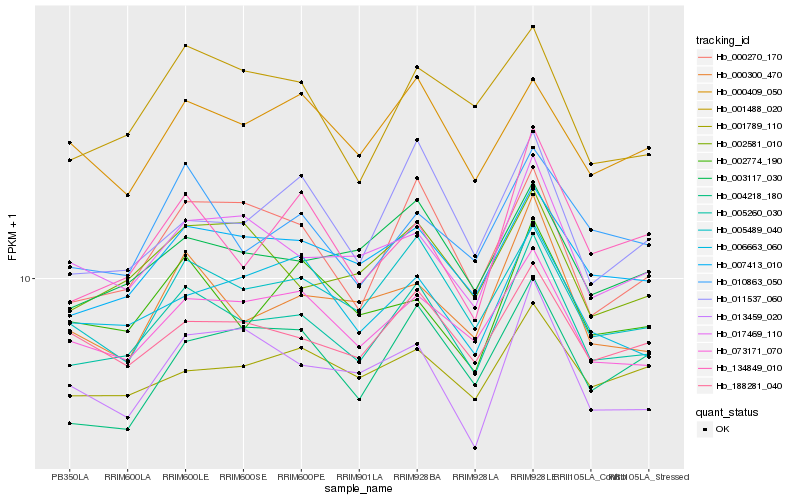
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005260_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 2 |
Hb_005489_040 |
0.0609813762 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 3 |
Hb_007413_010 |
0.0614385759 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 4 |
Hb_073171_070 |
0.0649641717 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 5 |
Hb_003117_030 |
0.0689807902 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 6 |
Hb_017469_110 |
0.0702736586 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 7 |
Hb_000270_170 |
0.0707700451 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 8 |
Hb_000300_470 |
0.0717844389 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 9 |
Hb_188281_040 |
0.0718182816 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 10 |
Hb_002581_010 |
0.0749150668 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 11 |
Hb_010863_050 |
0.0751389312 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 12 |
Hb_006663_060 |
0.0779212494 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 13 |
Hb_001488_020 |
0.0786925972 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 14 |
Hb_013459_020 |
0.0801094725 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 15 |
Hb_002774_190 |
0.0811200856 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 16 |
Hb_134849_010 |
0.0819713144 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 17 |
Hb_011537_060 |
0.0825948012 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 18 |
Hb_001789_110 |
0.0829789758 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_004218_180 |
0.0836795765 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 20 |
Hb_000409_050 |
0.0843771138 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |