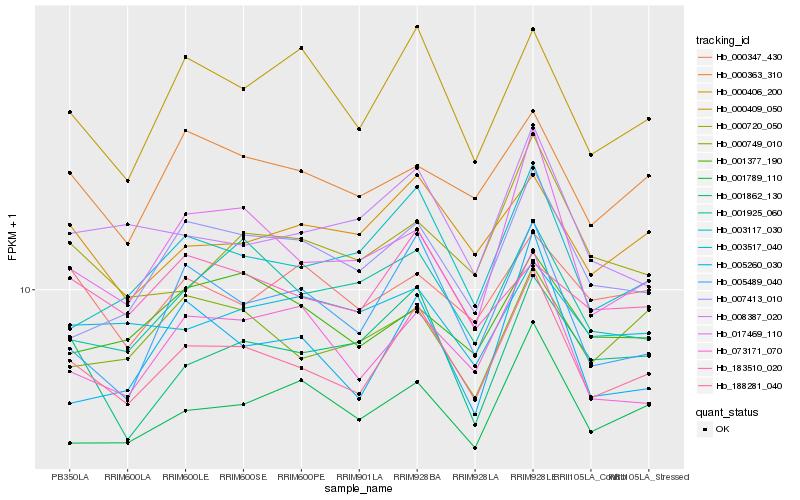
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_188281_040 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 2 |
Hb_001789_110 |
0.0609719363 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000406_200 |
0.0623046208 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 4 |
Hb_003117_030 |
0.0644700464 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 5 |
Hb_005489_040 |
0.0658712524 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 6 |
Hb_000409_050 |
0.0666893371 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 7 |
Hb_000363_310 |
0.0666905009 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 8 |
Hb_017469_110 |
0.0667692319 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 9 |
Hb_073171_070 |
0.0668581958 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 10 |
Hb_000720_050 |
0.0676465499 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001862_130 |
0.0689505356 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 12 |
Hb_008387_020 |
0.0696204379 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 13 |
Hb_000749_010 |
0.0712005349 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005260_030 |
0.0718182816 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 15 |
Hb_183510_020 |
0.0723500385 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 16 |
Hb_001377_190 |
0.0728838049 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_007413_010 |
0.0733189262 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 18 |
Hb_001925_060 |
0.0735319853 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 19 |
Hb_003517_040 |
0.0746577589 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000347_430 |
0.074902947 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |