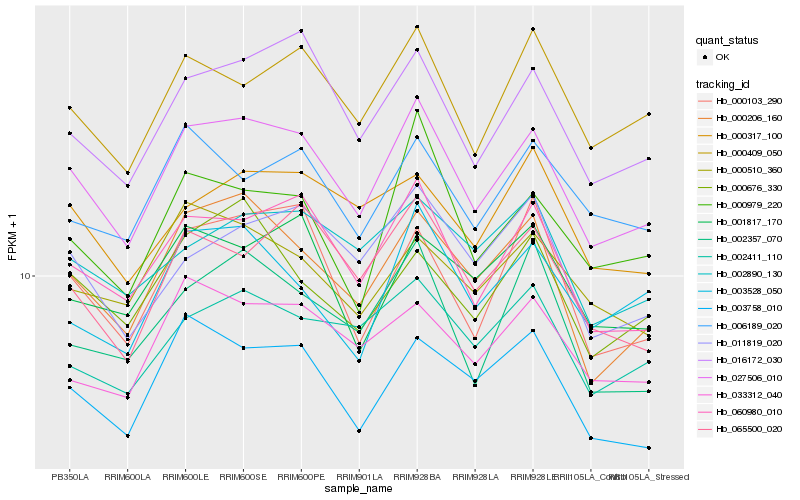
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027506_010 |
0.0 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 2 |
Hb_060980_010 |
0.0442334508 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_016172_030 |
0.0512219878 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 4 |
Hb_000206_160 |
0.0524035736 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_003758_010 |
0.0589880327 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000409_050 |
0.0597661427 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 7 |
Hb_065500_020 |
0.0642980181 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 8 |
Hb_000103_290 |
0.0649190376 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 9 |
Hb_000979_220 |
0.0650740487 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011819_020 |
0.0660293987 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 11 |
Hb_033312_040 |
0.0668502234 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000676_330 |
0.0669048854 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_002411_110 |
0.067149656 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 14 |
Hb_000510_360 |
0.0687357183 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 15 |
Hb_001817_170 |
0.0692889507 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 16 |
Hb_006189_020 |
0.0693013462 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 17 |
Hb_000317_100 |
0.0693189018 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 18 |
Hb_002357_070 |
0.0693343688 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 19 |
Hb_002890_130 |
0.0694380006 |
- |
- |
tip120, putative [Ricinus communis] |
| 20 |
Hb_003528_050 |
0.0705016766 |
- |
- |
protein transporter, putative [Ricinus communis] |