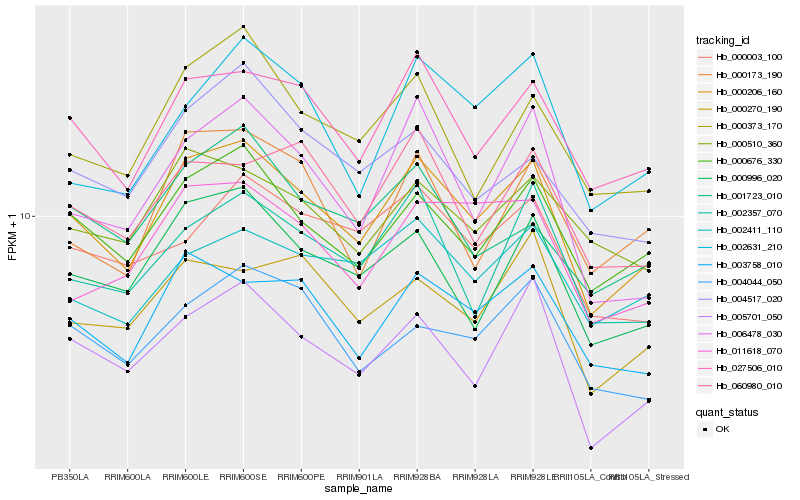
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000206_160 |
0.0 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_027506_010 |
0.0524035736 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 3 |
Hb_000676_330 |
0.0545265816 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_003758_010 |
0.0627536227 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005701_050 |
0.0642423824 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 6 |
Hb_000996_020 |
0.0657404524 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 7 |
Hb_000373_170 |
0.0743427934 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 8 |
Hb_002357_070 |
0.0751069173 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 9 |
Hb_002411_110 |
0.0752163206 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 10 |
Hb_000270_190 |
0.0756165731 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 11 |
Hb_006478_030 |
0.0758802624 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 12 |
Hb_004044_050 |
0.076076395 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 13 |
Hb_060980_010 |
0.0764672811 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_011618_070 |
0.0769590614 |
- |
- |
NMDA receptor-regulated protein [Medicago truncatula] |
| 15 |
Hb_000510_360 |
0.0770415554 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 16 |
Hb_002631_210 |
0.0780576711 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 17 |
Hb_000003_100 |
0.0780737635 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 18 |
Hb_001723_010 |
0.0790245269 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 19 |
Hb_004517_020 |
0.0790383486 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 20 |
Hb_000173_190 |
0.0791649067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |