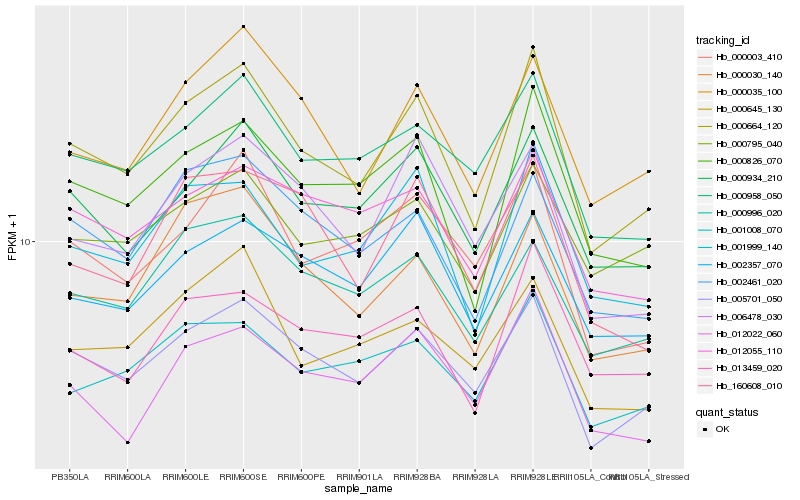
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000664_120 |
0.0 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005701_050 |
0.0531494693 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 3 |
Hb_000826_070 |
0.0625878706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001999_140 |
0.0658595911 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_000934_210 |
0.069028233 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 6 |
Hb_000035_100 |
0.0725626782 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_002461_020 |
0.0760460591 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000958_050 |
0.0761054435 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 9 |
Hb_000030_140 |
0.0765832686 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002357_070 |
0.078206435 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 11 |
Hb_001008_070 |
0.0803106694 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 12 |
Hb_013459_020 |
0.0805166809 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 13 |
Hb_000996_020 |
0.0810697188 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 14 |
Hb_000003_410 |
0.0836610302 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 15 |
Hb_006478_030 |
0.0840795113 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 16 |
Hb_012022_060 |
0.0841611497 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 17 |
Hb_012055_110 |
0.0848019636 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 18 |
Hb_000645_130 |
0.0862635525 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_000795_040 |
0.0875569398 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 20 |
Hb_160608_010 |
0.0875716164 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |