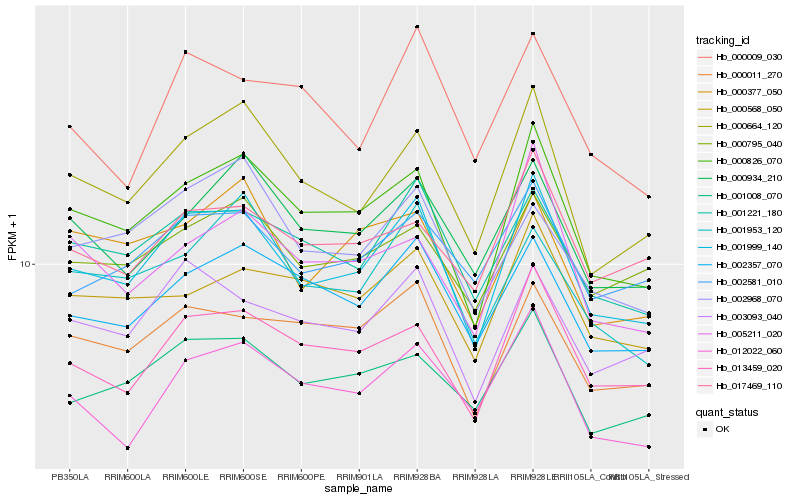
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_140 |
0.0 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_000826_070 |
0.0549352178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000664_120 |
0.0658595911 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_013459_020 |
0.071477713 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 5 |
Hb_001221_180 |
0.074074402 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 6 |
Hb_003093_040 |
0.0749513378 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_002357_070 |
0.0794187317 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 8 |
Hb_000011_270 |
0.0798475279 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 9 |
Hb_001008_070 |
0.0805368843 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 10 |
Hb_002581_010 |
0.0857093935 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000934_210 |
0.0868451223 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 12 |
Hb_012022_060 |
0.0873427496 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000568_050 |
0.0894222799 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000795_040 |
0.0905777054 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 15 |
Hb_017469_110 |
0.0923087106 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 16 |
Hb_001953_120 |
0.0925544169 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000377_050 |
0.094292076 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 18 |
Hb_000009_030 |
0.0955406495 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 19 |
Hb_005211_020 |
0.0967049867 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002968_070 |
0.0967939734 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |