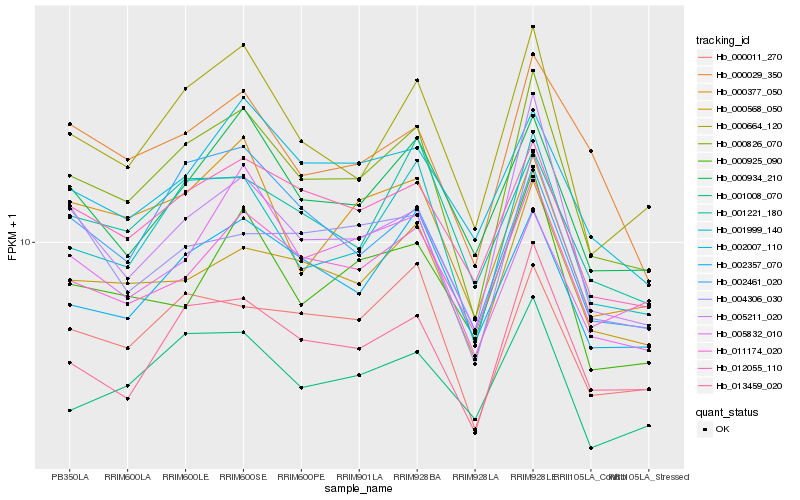
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000826_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001999_140 |
0.0549352178 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_013459_020 |
0.0620955615 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 4 |
Hb_000664_120 |
0.0625878706 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005211_020 |
0.069989394 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000568_050 |
0.0705306288 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_012055_110 |
0.071039614 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 8 |
Hb_000011_270 |
0.0733812043 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 9 |
Hb_000934_210 |
0.0779978818 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 10 |
Hb_002357_070 |
0.0794969269 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 11 |
Hb_011174_020 |
0.0814370835 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 12 |
Hb_000377_050 |
0.0825066557 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 13 |
Hb_002461_020 |
0.0829051978 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001008_070 |
0.0835639995 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 15 |
Hb_000029_350 |
0.0851187914 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 16 |
Hb_004306_030 |
0.0853142981 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 17 |
Hb_001221_180 |
0.0854585918 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 18 |
Hb_002007_110 |
0.0861712431 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000925_090 |
0.0862834443 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005832_010 |
0.0877117911 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |