| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
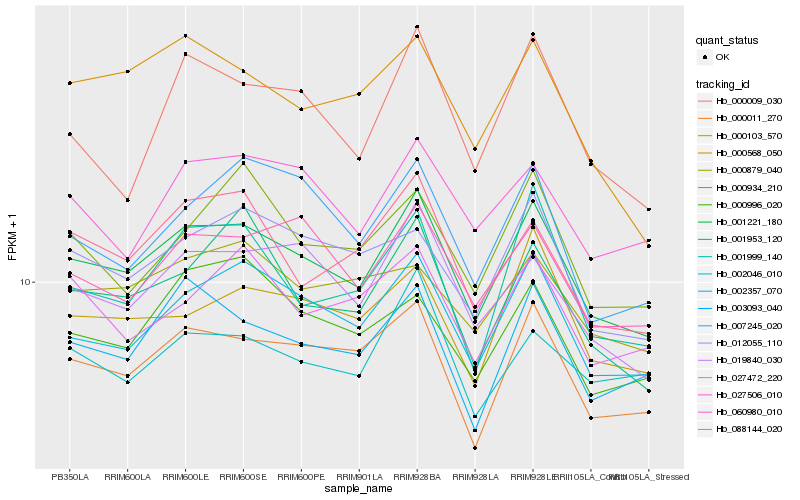
Hb_001221_180 |
0.0 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 2 |
Hb_002357_070 |
0.0529345033 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 3 |
Hb_000011_270 |
0.058193718 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 4 |
Hb_019840_030 |
0.0664058375 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 5 |
Hb_060980_010 |
0.0671813023 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_003093_040 |
0.0694246981 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_000568_050 |
0.0714676475 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_012055_110 |
0.0718624346 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 9 |
Hb_001999_140 |
0.074074402 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_088144_020 |
0.0743982706 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_027506_010 |
0.0758700861 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 12 |
Hb_000103_570 |
0.077090566 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001953_120 |
0.0777095268 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000009_030 |
0.079458741 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 15 |
Hb_027472_220 |
0.0796980673 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_007245_020 |
0.0799513781 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000879_040 |
0.0805752566 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 18 |
Hb_002046_010 |
0.0810222287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000934_210 |
0.0811559487 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 20 |
Hb_000996_020 |
0.0824049243 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |