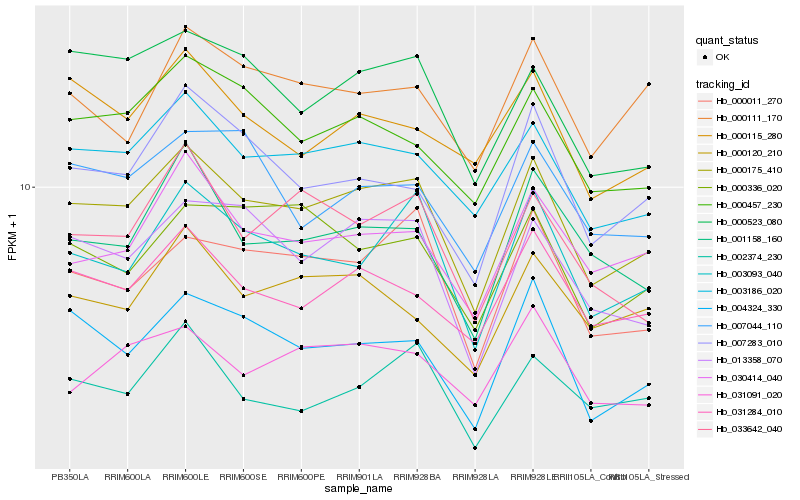
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_410 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_003186_020 |
0.053599513 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003093_040 |
0.062601556 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_013358_070 |
0.0662859488 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_031284_010 |
0.067636379 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 6 |
Hb_000011_270 |
0.0683598781 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 7 |
Hb_000523_080 |
0.0689979124 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000457_230 |
0.0706093069 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_030414_040 |
0.0716844638 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004324_330 |
0.0740394143 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 11 |
Hb_007283_010 |
0.0743789211 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 12 |
Hb_001158_160 |
0.0770958424 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_033642_040 |
0.0782271279 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000336_020 |
0.0788317354 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 15 |
Hb_000120_210 |
0.0797894038 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 16 |
Hb_031091_020 |
0.0813611881 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 17 |
Hb_002374_230 |
0.0817379648 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 18 |
Hb_000111_170 |
0.08233006 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007044_110 |
0.0827119351 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000115_280 |
0.0828550139 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |