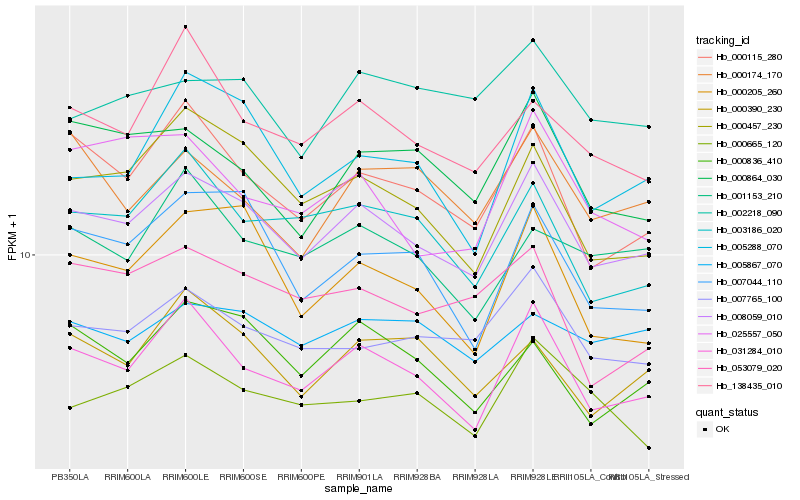
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008059_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 2 |
Hb_031284_010 |
0.0410615737 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 3 |
Hb_007765_100 |
0.0565267476 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 4 |
Hb_000115_280 |
0.0587168422 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 5 |
Hb_000457_230 |
0.061569753 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000836_410 |
0.0651291222 |
- |
- |
sec10, putative [Ricinus communis] |
| 7 |
Hb_000205_260 |
0.0659984756 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 8 |
Hb_003186_020 |
0.0662821766 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007044_110 |
0.0665264027 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_005288_070 |
0.0669777899 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000390_230 |
0.0671706035 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 12 |
Hb_000864_030 |
0.0673767773 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000665_120 |
0.0676312561 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 14 |
Hb_138435_010 |
0.0677718509 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 15 |
Hb_005867_070 |
0.0692063552 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_001153_210 |
0.0696596293 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002218_090 |
0.0711824603 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 18 |
Hb_053079_020 |
0.0716803521 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 19 |
Hb_025557_050 |
0.072293991 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 20 |
Hb_000174_170 |
0.0729940782 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |