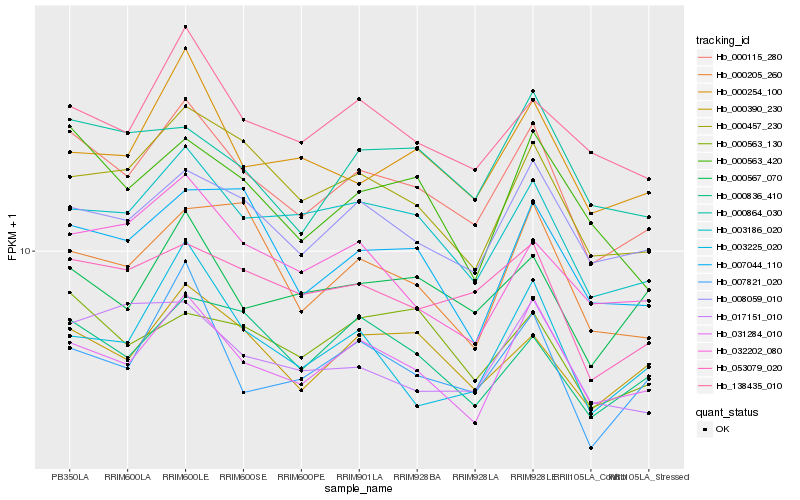
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_280 |
0.0 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 2 |
Hb_031284_010 |
0.0490709682 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 3 |
Hb_003186_020 |
0.0562598408 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008059_010 |
0.0587168422 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000390_230 |
0.0607048345 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 6 |
Hb_053079_020 |
0.0616401323 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 7 |
Hb_000457_230 |
0.0643121593 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_138435_010 |
0.0662545327 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 9 |
Hb_000836_410 |
0.0673253834 |
- |
- |
sec10, putative [Ricinus communis] |
| 10 |
Hb_000567_070 |
0.0689344332 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 11 |
Hb_007821_020 |
0.0695623447 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000864_030 |
0.0706115346 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000563_130 |
0.0716461521 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 14 |
Hb_000254_100 |
0.0721031307 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 15 |
Hb_000563_420 |
0.0724068582 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 16 |
Hb_017151_010 |
0.0732899205 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 17 |
Hb_007044_110 |
0.0753909647 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_003225_020 |
0.0771117319 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 19 |
Hb_032202_080 |
0.0773170467 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000205_260 |
0.0779254883 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |