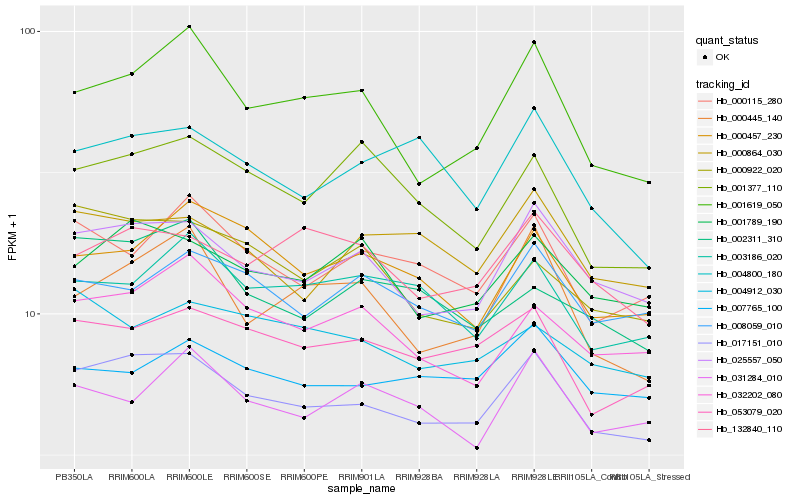
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017151_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 2 |
Hb_025557_050 |
0.0473575065 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 3 |
Hb_053079_020 |
0.0682492314 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 4 |
Hb_001789_190 |
0.06930241 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 5 |
Hb_000115_280 |
0.0732899205 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 6 |
Hb_001619_050 |
0.0744289836 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 7 |
Hb_032202_080 |
0.0756657483 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000864_030 |
0.0776825705 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_008059_010 |
0.0788965649 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 10 |
Hb_132840_110 |
0.0789002262 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000457_230 |
0.081254301 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_007765_100 |
0.0812730248 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 13 |
Hb_004912_030 |
0.0821805163 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 14 |
Hb_003186_020 |
0.0828501335 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 15 |
Hb_031284_010 |
0.0834794226 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 16 |
Hb_004800_180 |
0.0845842875 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 17 |
Hb_002311_310 |
0.0855441933 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 18 |
Hb_000445_140 |
0.0856365669 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 19 |
Hb_000922_020 |
0.0869619618 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_001377_110 |
0.0874847335 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |