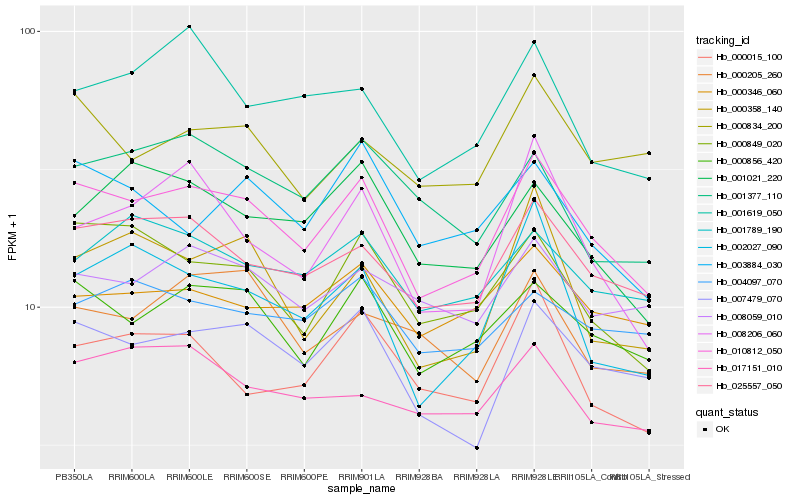
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010812_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000856_420 |
0.0659206738 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_025557_050 |
0.0696091871 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 4 |
Hb_007479_070 |
0.0715265921 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 5 |
Hb_000849_020 |
0.0797983979 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001789_190 |
0.0917087837 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 7 |
Hb_002027_090 |
0.0938068148 |
- |
- |
- |
| 8 |
Hb_008059_010 |
0.0945290434 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001021_220 |
0.095137572 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000015_100 |
0.0953019707 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_008206_060 |
0.0956795165 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000358_140 |
0.0959554436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000346_060 |
0.0966021124 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 14 |
Hb_003884_030 |
0.0966383075 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 15 |
Hb_001619_050 |
0.0968085343 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 16 |
Hb_017151_010 |
0.0985826676 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 17 |
Hb_000834_200 |
0.0989520272 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 18 |
Hb_004097_070 |
0.099779725 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 19 |
Hb_001377_110 |
0.1001851523 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 20 |
Hb_000205_260 |
0.1011269552 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |