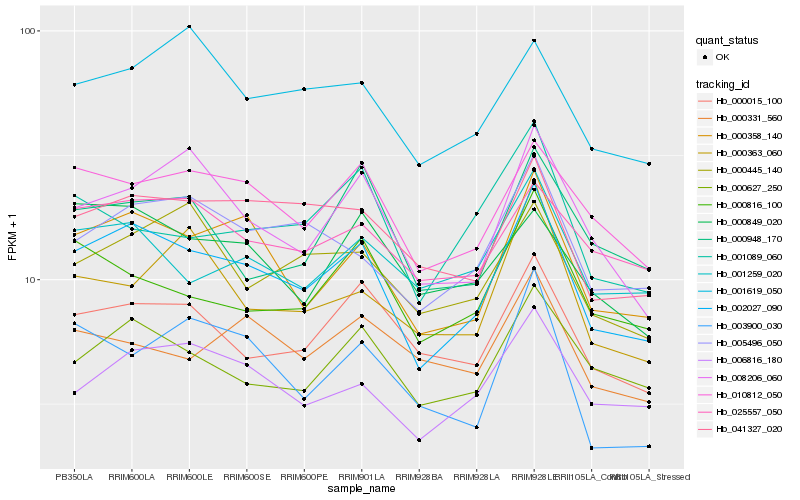
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000358_140 |
0.0699665875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_010812_050 |
0.0938068148 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005496_050 |
0.0968764912 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 5 |
Hb_041327_020 |
0.0969588818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006816_180 |
0.0970859458 |
- |
- |
- |
| 7 |
Hb_000627_250 |
0.0983373762 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000015_100 |
0.0992507514 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_008206_060 |
0.0997682781 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001259_020 |
0.1077049647 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000816_100 |
0.1088284767 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_025557_050 |
0.1090902332 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 13 |
Hb_001619_050 |
0.1126588932 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000445_140 |
0.1131616465 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 15 |
Hb_000849_020 |
0.1137321978 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000948_170 |
0.1162631999 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 17 |
Hb_001089_060 |
0.1193481541 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1A isoform X1 [Jatropha curcas] |
| 18 |
Hb_000331_560 |
0.1225967614 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 19 |
Hb_000363_060 |
0.1227741202 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003900_030 |
0.1228437095 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |