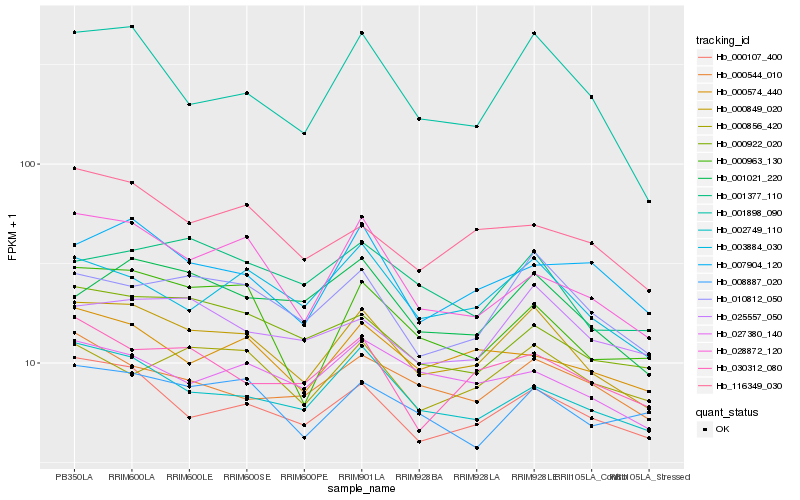
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000849_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000107_400 |
0.0773809177 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870-like [Jatropha curcas] |
| 3 |
Hb_010812_050 |
0.0797983979 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003884_030 |
0.0832187581 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 5 |
Hb_002749_110 |
0.0834522718 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001898_090 |
0.0885018893 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001021_220 |
0.0936858812 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000963_130 |
0.0945119698 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_028872_120 |
0.0954918116 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 10 |
Hb_000922_020 |
0.0955869592 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_025557_050 |
0.0968679512 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 12 |
Hb_000574_440 |
0.0972086694 |
- |
- |
PREDICTED: uncharacterized protein LOC105634833 [Jatropha curcas] |
| 13 |
Hb_000856_420 |
0.0977227616 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_030312_080 |
0.0986129948 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 15 |
Hb_001377_110 |
0.0987752905 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 16 |
Hb_008887_020 |
0.0988072295 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 17 |
Hb_027380_140 |
0.0991854243 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 18 |
Hb_116349_030 |
0.0995423278 |
- |
- |
PREDICTED: zinc finger MYND domain-containing protein 15 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000544_010 |
0.099772634 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_007904_120 |
0.0999496173 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |