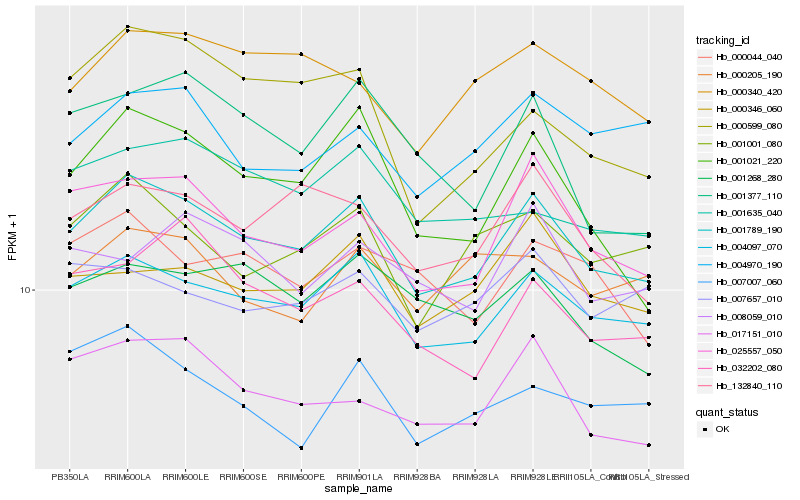
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 2 |
Hb_004097_070 |
0.0380910407 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 3 |
Hb_025557_050 |
0.0573874558 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 4 |
Hb_132840_110 |
0.067770742 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_017151_010 |
0.06930241 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 6 |
Hb_001001_080 |
0.0695034685 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 7 |
Hb_001021_220 |
0.0697066085 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000346_060 |
0.0732509001 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 9 |
Hb_001268_280 |
0.0744011845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001635_040 |
0.0746791272 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 11 |
Hb_000340_420 |
0.0758285424 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_000044_040 |
0.0778532783 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 13 |
Hb_032202_080 |
0.079014146 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_007007_060 |
0.0790155465 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004970_190 |
0.0805950037 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_008059_010 |
0.0808312581 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007657_010 |
0.0816287637 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_001377_110 |
0.0830308573 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 19 |
Hb_000205_190 |
0.0835281378 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000599_080 |
0.0850703419 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |