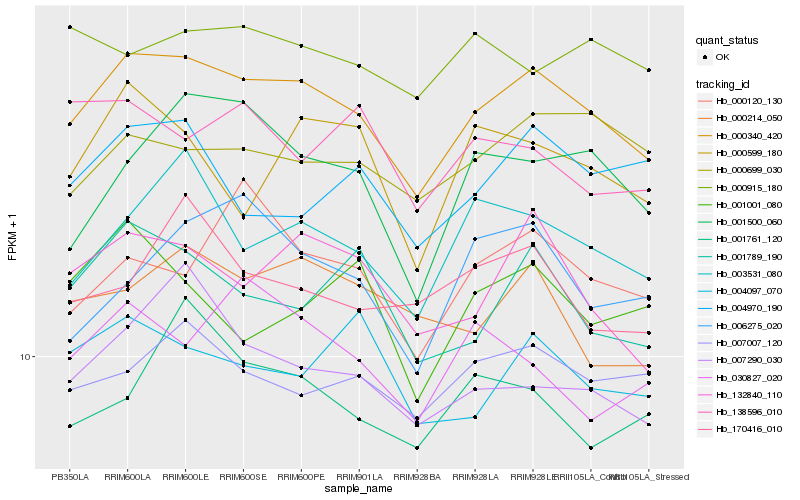
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_420 |
0.0 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_132840_110 |
0.0689004163 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_003531_080 |
0.0702627265 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007007_120 |
0.0740587121 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001789_190 |
0.0758285424 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 6 |
Hb_007290_030 |
0.0794925876 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 7 |
Hb_000214_050 |
0.0797586475 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000699_030 |
0.0809520629 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001500_060 |
0.0810000476 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein ZFN-like [Jatropha curcas] |
| 10 |
Hb_004097_070 |
0.0828096039 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 11 |
Hb_000120_130 |
0.0835242358 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 12 |
Hb_000599_180 |
0.0835935477 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_170416_010 |
0.0851290373 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 14 |
Hb_000915_180 |
0.0864973574 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Pyrus x bretschneideri] |
| 15 |
Hb_004970_190 |
0.0867961152 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_006275_020 |
0.087992317 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 17 |
Hb_030827_020 |
0.0901299089 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 18 |
Hb_001761_120 |
0.090237248 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 19 |
Hb_001001_080 |
0.0903947063 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 20 |
Hb_138596_010 |
0.0904340103 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |