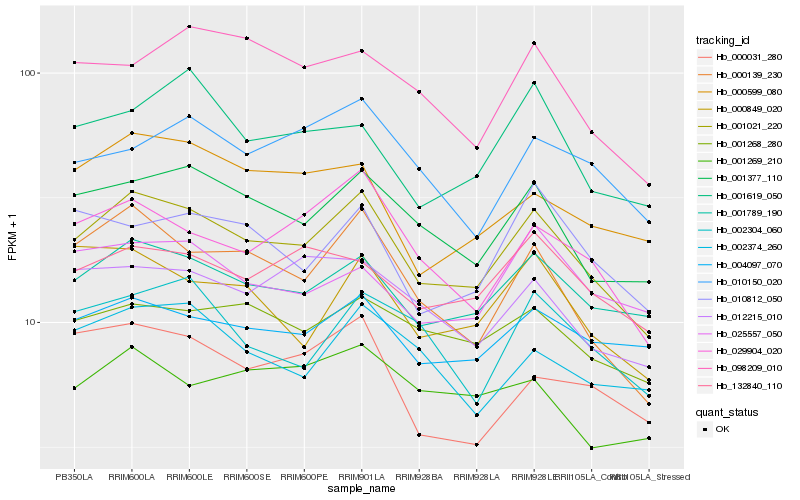
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001021_220 |
0.0 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001377_110 |
0.069054735 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 3 |
Hb_001789_190 |
0.0697066085 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 4 |
Hb_000139_230 |
0.0784891678 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 5 |
Hb_029904_020 |
0.079950613 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 6 |
Hb_001268_280 |
0.0867504466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_132840_110 |
0.0895314636 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_004097_070 |
0.0900157221 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 9 |
Hb_012215_010 |
0.0904997518 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 10 |
Hb_000599_080 |
0.0931357213 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 11 |
Hb_000849_020 |
0.0936858812 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002374_260 |
0.0950724359 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 13 |
Hb_010812_050 |
0.095137572 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 14 |
Hb_098209_010 |
0.0964606992 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 15 |
Hb_025557_050 |
0.0965535083 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 16 |
Hb_002304_060 |
0.0967806943 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 17 |
Hb_010150_020 |
0.0971066485 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 18 |
Hb_001619_050 |
0.0985083943 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001269_210 |
0.099253219 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000031_280 |
0.0998372028 |
- |
- |
- |