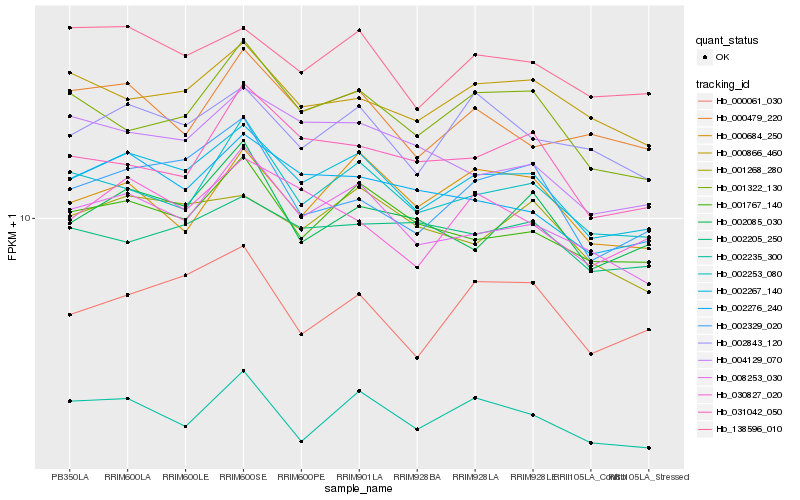
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002267_140 |
0.0 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 2 |
Hb_008253_030 |
0.0480598746 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 3 |
Hb_001767_140 |
0.0527616533 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 4 |
Hb_138596_010 |
0.0555505684 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 5 |
Hb_002276_240 |
0.0557169968 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 6 |
Hb_002253_080 |
0.0557951078 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 7 |
Hb_002843_120 |
0.0587323478 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001268_280 |
0.0616608224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001322_130 |
0.0630998465 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 10 |
Hb_002329_020 |
0.0632006784 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 11 |
Hb_000866_460 |
0.0641614691 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_030827_020 |
0.0642634956 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 13 |
Hb_000684_250 |
0.0656834397 |
- |
- |
PREDICTED: uncharacterized protein LOC105642172 [Jatropha curcas] |
| 14 |
Hb_002235_300 |
0.0662664305 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000479_220 |
0.0678151448 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 16 |
Hb_000061_030 |
0.0679221908 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065 [Jatropha curcas] |
| 17 |
Hb_031042_050 |
0.0692925796 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 18 |
Hb_002205_250 |
0.069361574 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002085_030 |
0.069535273 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004129_070 |
0.0701134074 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |