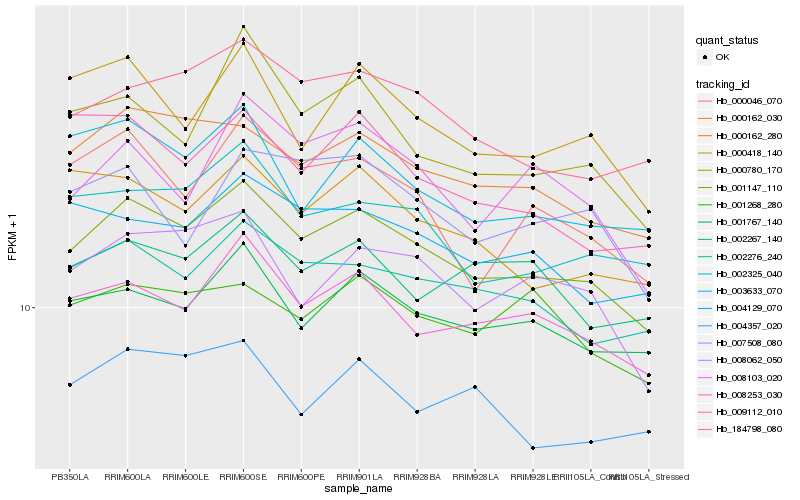
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001147_110 |
0.0 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 2 |
Hb_000162_030 |
0.0545624809 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_008253_030 |
0.0565024109 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 4 |
Hb_002276_240 |
0.0571270997 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 5 |
Hb_001767_140 |
0.0605042247 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 6 |
Hb_184798_080 |
0.0622469184 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 7 |
Hb_002325_040 |
0.0657678593 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008062_050 |
0.0666290534 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 9 |
Hb_004129_070 |
0.0696434973 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 10 |
Hb_000418_140 |
0.0699795653 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 11 |
Hb_003633_070 |
0.0709437665 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 12 |
Hb_000162_280 |
0.0720288238 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 13 |
Hb_000046_070 |
0.0721956985 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000780_170 |
0.0723051218 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 15 |
Hb_001268_280 |
0.0755508349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002267_140 |
0.0756923571 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 17 |
Hb_004357_020 |
0.0777584023 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 18 |
Hb_008103_020 |
0.0785231417 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 19 |
Hb_009112_010 |
0.0792255222 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 20 |
Hb_007508_080 |
0.0797317002 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |