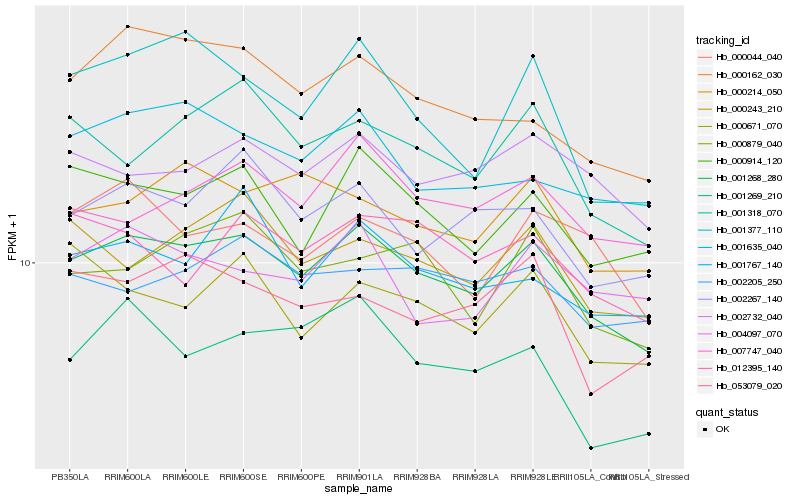
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001268_280 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000914_120 |
0.0600866545 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002267_140 |
0.0616608224 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 4 |
Hb_000162_030 |
0.0639331467 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_001318_070 |
0.0641855502 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002732_040 |
0.0642103171 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000243_210 |
0.0652776878 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_001635_040 |
0.0666474006 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 9 |
Hb_000879_040 |
0.0667691876 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 10 |
Hb_007747_040 |
0.0672236082 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 11 |
Hb_000214_050 |
0.0677330193 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000044_040 |
0.067780089 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 13 |
Hb_001767_140 |
0.0684429549 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 14 |
Hb_012395_140 |
0.0689732037 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 15 |
Hb_002205_250 |
0.0697848437 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001377_110 |
0.0705704082 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 17 |
Hb_001269_210 |
0.0718008509 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000671_070 |
0.0725352529 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 19 |
Hb_053079_020 |
0.0727581364 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 20 |
Hb_004097_070 |
0.0729041668 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |