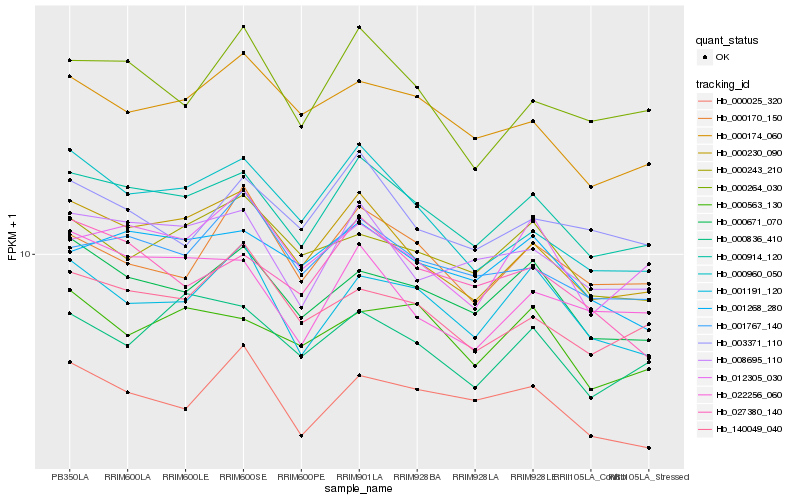
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000914_120 |
0.0 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000230_090 |
0.0463163406 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008695_110 |
0.0520012017 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000671_070 |
0.0543482351 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 5 |
Hb_001268_280 |
0.0600866545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000264_030 |
0.0618589032 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 7 |
Hb_001767_140 |
0.0629475612 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 8 |
Hb_000174_060 |
0.0639052982 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 9 |
Hb_001191_120 |
0.0643302849 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 10 |
Hb_140049_040 |
0.0648129022 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 11 |
Hb_000960_050 |
0.0649697382 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 12 |
Hb_003371_110 |
0.0654626359 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 13 |
Hb_000836_410 |
0.0656446267 |
- |
- |
sec10, putative [Ricinus communis] |
| 14 |
Hb_012305_030 |
0.0664606006 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 15 |
Hb_000563_130 |
0.0669526126 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 16 |
Hb_027380_140 |
0.0673761737 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 17 |
Hb_022256_060 |
0.0674851536 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 18 |
Hb_000170_150 |
0.068950426 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000243_210 |
0.0701450381 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000025_320 |
0.0702090306 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |